4XIA
 
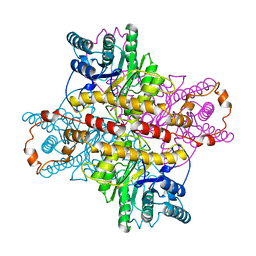 | | STRUCTURES OF D-XYLOSE ISOMERASE FROM ARTHROBACTER STRAIN B3728 CONTAINING THE INHIBITORS XYLITOL AND D-SORBITOL AT 2.5 ANGSTROMS AND 2.3 ANGSTROMS RESOLUTION, RESPECTIVELY | | Descriptor: | D-XYLOSE ISOMERASE, MAGNESIUM ION, sorbitol | | Authors: | Henrick, K, Collyer, C.A, Blow, D.M. | | Deposit date: | 1989-07-05 | | Release date: | 1990-04-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of D-xylose isomerase from Arthrobacter strain B3728 containing the inhibitors xylitol and D-sorbitol at 2.5 A and 2.3 A resolution, respectively.
J.Mol.Biol., 208, 1989
|
|
3BSB
 
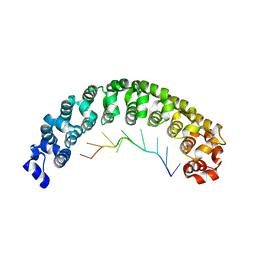 | | Crystal Structure of Human Pumilio1 in Complex with CyclinB reverse RNA | | Descriptor: | 5'-R(*UP*UP*UP*AP*AP*UP*GP*UP*U)-3', Pumilio homolog 1 | | Authors: | Gupta, Y.K, Nair, D.T, Wharton, R.P, Aggarwal, A.K. | | Deposit date: | 2007-12-23 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity.
Structure, 16, 2008
|
|
5B4X
 
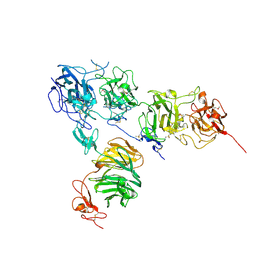 | | Crystal structure of the ApoER2 ectodomain in complex with the Reelin R56 fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Low density lipoprotein receptor-related protein 8, ... | | Authors: | Yasui, N, Hirai, H, Yamashita, K, Takagi, J, Nogi, T. | | Deposit date: | 2016-04-20 | | Release date: | 2017-04-26 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Crystal structure of the ectodomain from a LDLR close homologue in complex with its physiological ligand.
To Be Published
|
|
5BNN
 
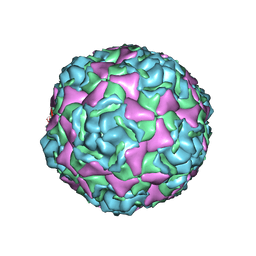 | | Crystal structure of human enterovirus D68 in complex with 6'SL | | Descriptor: | Capsid protein VP1, Capsid protein VP2, Capsid protein VP3, ... | | Authors: | Liu, Y, Sheng, J, Meng, G, Xiao, C, Rossmann, M.G. | | Deposit date: | 2015-05-26 | | Release date: | 2015-11-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Sialic acid-dependent cell entry of human enterovirus D68.
Nat Commun, 6, 2015
|
|
3BXN
 
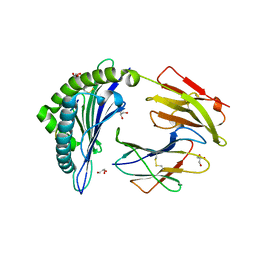 | | The high resolution crystal structure of HLA-B*1402 complexed with a Cathepsin A signal sequence peptide, pCatA | | Descriptor: | Cathepsin A signal sequence octapeptide, GLYCEROL, HLA-B*1402 extracellular domain, ... | | Authors: | Kumar, P, Vahedi-Faridi, A, Saenger, W, Uchanska-Ziegler, B, Ziegler, A. | | Deposit date: | 2008-01-14 | | Release date: | 2009-02-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.864 Å) | | Cite: | Structural basis for T cell alloreactivity among three HLA-B14 and HLA-B27 antigens
J.Biol.Chem., 284, 2009
|
|
3C76
 
 | |
4HP2
 
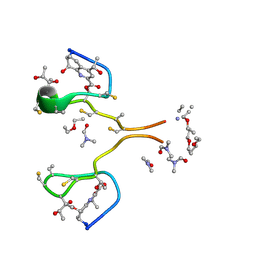 | | Invariom refinement of a new dimeric monoclinic 2 solvate of thiostrepton at 0.64 angstrom resolution | | Descriptor: | DIMETHYLFORMAMIDE, Thiostrepton, diethyl ether | | Authors: | Proepper, K, Holstein, J.J, Huebschle, C.B, Bond, C.S, Dittrich, B. | | Deposit date: | 2012-10-23 | | Release date: | 2013-10-02 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (0.64 Å) | | Cite: | Invariom refinement of a new monoclinic solvate of thiostrepton at 0.64 angstrom resolution.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
3CBR
 
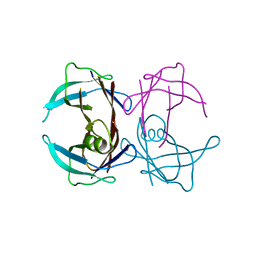 | | Crystal structure of human Transthyretin (TTR) at pH3.5 | | Descriptor: | Transthyretin | | Authors: | Mohamedmohaideen, N.N, Palaninathan, S.K, Snee, W.C, Kelly, J.W, C Sacchettini, J. | | Deposit date: | 2008-02-22 | | Release date: | 2008-08-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural insight into pH-induced conformational changes within the native human transthyretin tetramer.
J.Mol.Biol., 382, 2008
|
|
4I3J
 
 | | Structures of PR1 intermediate of photoactive yellow protein E46Q mutant from time-resolved laue crystallography collected AT 14ID APS | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, Photoactive yellow protein | | Authors: | Jung, Y.O, Lee, J.H, Kim, J, Schmidt, M, Vukica, S, Moffat, K, Ihee, H. | | Deposit date: | 2012-11-26 | | Release date: | 2013-03-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Volume-conserving trans-cis isomerization pathways in photoactive yellow protein visualized by picosecond X-ray crystallography
NAT.CHEM., 5, 2013
|
|
4XSZ
 
 | | Crystal structure of CBR 9393 bound to Escherichia coli RNA polymerase holoenzyme | | Descriptor: | 4-[3-(4-fluorophenyl)-1H-pyrazol-4-yl]-N-[2-(piperazin-1-yl)ethyl]-2-(trifluoromethyl)aniline, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2015-01-22 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.683 Å) | | Cite: | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5CGI
 
 | | Yeast 20S proteasome beta5-G48C mutant in complex with ONX 0914 | | Descriptor: | 1,2,4-trideoxy-4-methyl-2-{[N-(morpholin-4-ylacetyl)-L-alanyl-O-methyl-L-tyrosyl]amino}-1-phenyl-D-xylitol, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Dubiella, C, Groll, M. | | Deposit date: | 2015-07-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective Inhibition of the Immunoproteasome by Structure-Based Targeting of a Non-catalytic Cysteine.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4IK5
 
 | | High resolution structure of Delta-REST-GCaMP3 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4F21
 
 | | Crystal structure of carboxylesterase/phospholipase family protein from Francisella tularensis | | Descriptor: | Carboxylesterase/phospholipase family protein, N-((1R,2S)-2-allyl-4-oxocyclobutyl)-4-methylbenzenesulfonamide, bound form | | Authors: | Filippova, E.V, Minasov, G, Kuhn, M, Wawrzak, Z, Shuvalova, L, Dubrovska, I, Winsor, J.R, Kiryukhina, O, Becker, D.P, Armoush, N, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-07 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Large scale structural rearrangement of a serine hydrolase from Francisella tularensis facilitates catalysis.
J.Biol.Chem., 288, 2013
|
|
4II4
 
 | |
5C0U
 
 | | Crystal structure of the copper-bound form of MerB mutant D99S | | Descriptor: | Alkylmercury lyase, BROMIDE ION, COPPER (II) ION | | Authors: | Wahba, H.M, Lecoq, L, Stevenson, M, Mansour, A, Cappadocia, L, Lafrance-Vanasse, J, Wilkinson, K.J, Sygusch, J, Wilcox, D.E, Omichinski, J.G. | | Deposit date: | 2015-06-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural and Biochemical Characterization of a Copper-Binding Mutant of the Organomercurial Lyase MerB: Insight into the Key Role of the Active Site Aspartic Acid in Hg-Carbon Bond Cleavage and Metal Binding Specificity.
Biochemistry, 55, 2016
|
|
4XSX
 
 | | Crystal structure of CBR 703 bound to Escherichia coli RNA polymerase holoenzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Bae, B, Darst, S.A. | | Deposit date: | 2015-01-22 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.708 Å) | | Cite: | CBR antimicrobials inhibit RNA polymerase via at least two bridge-helix cap-mediated effects on nucleotide addition.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4IK1
 
 | | High resolution structure of GCaMPJ at pH 8.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
3BSX
 
 | | Crystal Structure of Human Pumilio 1 in complex with Puf5 RNA | | Descriptor: | 5'-R(*UP*UP*GP*UP*AP*AP*UP*AP*UP*UP*A)-3', Pumilio homolog 1 | | Authors: | Gupta, Y.K, Nair, D.T, Wharton, R.P, Aggarwal, A.K. | | Deposit date: | 2007-12-26 | | Release date: | 2008-04-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Structures of human Pumilio with noncognate RNAs reveal molecular mechanisms for binding promiscuity.
Structure, 16, 2008
|
|
4F74
 
 | |
3CCV
 
 | |
3CC7
 
 | |
5CGY
 
 | |
4IHF
 
 | | Chasing Acyl Carrier Protein Through a Catalytic Cycle of Lipid A Production | | Descriptor: | Acyl carrier protein, CHLORIDE ION, S-[2-({N-[(2S)-2-hydroxy-3,3-dimethyl-4-(phosphonooxy)butanoyl]-beta-alanyl}amino)ethyl] (3R)-3-hydroxytetradecanethioate, ... | | Authors: | Masoudi, A, Raetz, C.R.H, Pemble, C.W. | | Deposit date: | 2012-12-18 | | Release date: | 2013-11-13 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Chasing acyl carrier protein through a catalytic cycle of lipid A production.
Nature, 505, 2014
|
|
5CHW
 
 | |
4IK4
 
 | | High resolution structure of GCaMP3 at pH 5.0 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
