1NB0
 
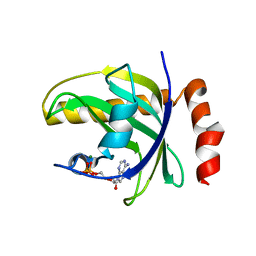 | | Crystal Structure of Human Riboflavin Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, hypothetical protein FLJ11149 | | Authors: | Karthikeyan, S, Zhou, Q, Mseeh, F, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-12-01 | | Release date: | 2003-03-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Riboflavin Kinase Reveals a Beta Barrel Fold and a Novel Active Site Arch
Structure, 11, 2003
|
|
1NB1
 
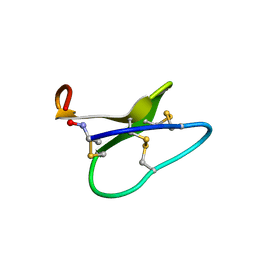 | | High resolution solution structure of kalata B1 | | Descriptor: | kalata B1 | | Authors: | Rosengren, K.J, Daly, N.L, Plan, M.R, Waine, C, Craik, D.J. | | Deposit date: | 2002-12-01 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Twists, Knots, and Rings in Proteins. STRUCTURAL DEFINITION OF THE CYCLOTIDE FRAMEWORK
J.Biol.Chem., 278, 2003
|
|
1NB2
 
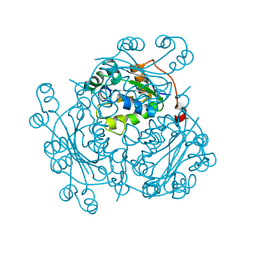 | | Crystal Structure of Nucleoside Diphosphate Kinase from Bacillus Halodenitrificans | | Descriptor: | Nucleoside Diphosphate Kinase | | Authors: | Chen, C.-J, Liu, M.-Y, Chang, W.-C, Chang, T, Wang, B.-C, Le Gall, J. | | Deposit date: | 2002-12-02 | | Release date: | 2003-05-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a nucleoside diphosphate kinase from Bacillus halodenitrificans: coexpression of its activity with a Mn-superoxide dismutase.
J.Struct.Biol., 142, 2003
|
|
1NB3
 
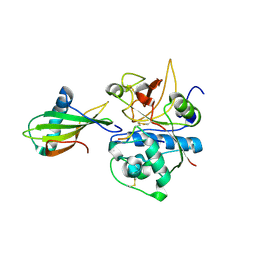 | | Crystal structure of stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo-and exopeptidases | | Descriptor: | CATHEPSIN H MINI CHAIN, Cathepsin H, Stefin A, ... | | Authors: | Jenko, S, Dolenc, I, Guncar, G, Dobersek, A, Podobnik, M, Turk, D. | | Deposit date: | 2002-12-02 | | Release date: | 2003-02-18 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo- and exopeptidases
J.Mol.Biol., 326, 2003
|
|
1NB4
 
 | |
1NB5
 
 | | Crystal structure of stefin A in complex with cathepsin H | | Descriptor: | Cathepsin H, Cathepsin H MINI CHAIN, STEFIN A, ... | | Authors: | Jenko, S, Dolenc, I, Guncar, G, Dobersek, A, Podobnik, M, Turk, D. | | Deposit date: | 2002-12-02 | | Release date: | 2003-02-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo- and exopeptidases
J.Mol.Biol., 326, 2003
|
|
1NB6
 
 | | HC-J4 RNA polymerase complexed with UTP | | Descriptor: | MANGANESE (II) ION, URIDINE 5'-TRIPHOSPHATE, polyprotein | | Authors: | O'Farrell, D.J, Trowbridge, R, Rowlands, D.J, Jaeger, J. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation.
J.Mol.Biol., 326, 2003
|
|
1NB7
 
 | | HC-J4 RNA polymerase complexed with short RNA template strand | | Descriptor: | 5'-R(*UP*UP*UP*U)-3', MANGANESE (II) ION, polyprotein | | Authors: | O'Farrell, D.J, Trowbridge, R, Rowlands, D.J, Jaeger, J. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Substrate complexes of hepatitis C virus RNA polymerase (HC-J4): structural evidence for nucleotide import and de-novo initiation.
J.Mol.Biol., 326, 2003
|
|
1NB8
 
 | | Structure of the catalytic domain of USP7 (HAUSP) | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Li, P, Li, M, Li, W, Yao, T, Wu, J.-W, Gu, W, Cohen, R.E, Shi, Y. | | Deposit date: | 2002-12-02 | | Release date: | 2003-01-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde
Cell(Cambridge,Mass.), 111, 2002
|
|
1NB9
 
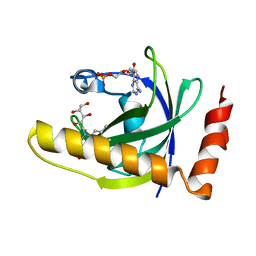 | | Crystal Structure of Riboflavin Kinase | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, RIBOFLAVIN, ... | | Authors: | Karthikeyan, S, Zhou, Q, Mseeh, F, Grishin, N.V, Osterman, A.L, Zhang, H. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-11 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of Human Riboflavin Kinase Reveals a Beta Barrel Fold and a Novel Active Site Arch
Structure, 11, 2003
|
|
1NBA
 
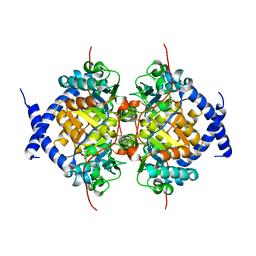 | | CRYSTAL STRUCTURE ANALYSIS, REFINEMENT AND ENZYMATIC REACTION MECHANISM OF N-CARBAMOYLSARCOSINE AMIDOHYDROLASE FROM ARTHROBACTER SP. AT 2.0 ANGSTROMS RESOLUTION | | Descriptor: | N-CARBAMOYLSARCOSINE AMIDOHYDROLASE, SULFATE ION | | Authors: | Romao, M.J, Turk, D, Gomis-Ruth, F.-Z, Huber, R, Schumacher, G, Mollering, H, Russmann, L. | | Deposit date: | 1992-05-18 | | Release date: | 1994-06-22 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure analysis, refinement and enzymatic reaction mechanism of N-carbamoylsarcosine amidohydrolase from Arthrobacter sp. at 2.0 A resolution.
J.Mol.Biol., 226, 1992
|
|
1NBB
 
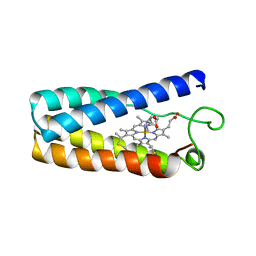 | | N-BUTYLISOCYANIDE BOUND RHODOBACTER CAPSULATUS CYTOCHROME C' | | Descriptor: | CYTOCHROME C', N-BUTYL ISOCYANIDE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tahirov, T.H, Misaki, S, Meyer, T.E, Cusanovich, M.A, Higuchi, Y, Yasuoka, N. | | Deposit date: | 1996-03-18 | | Release date: | 1996-08-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Concerted movement of side chains in the haem vicinity observed on ligand binding in cytochrome c' from rhodobacter capsulatus.
Nat.Struct.Biol., 3, 1996
|
|
1NBC
 
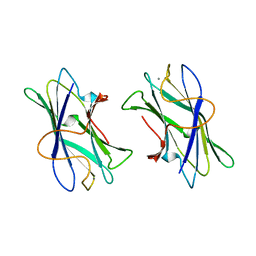 | | BACTERIAL TYPE 3A CELLULOSE-BINDING DOMAIN | | Descriptor: | CALCIUM ION, CELLULOSOMAL SCAFFOLDING PROTEIN A | | Authors: | Tormo, J, Lamed, R, Steitz, T.A. | | Deposit date: | 1996-09-10 | | Release date: | 1997-09-26 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a bacterial family-III cellulose-binding domain: a general mechanism for attachment to cellulose.
EMBO J., 15, 1996
|
|
1NBE
 
 | |
1NBF
 
 | | Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde | | Descriptor: | Ubiquitin aldehyde, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Hu, M, Li, P, Li, M, Li, W, Yao, T, Wu, J.-W, Gu, W, Cohen, R.E, Shi, Y. | | Deposit date: | 2002-12-02 | | Release date: | 2003-01-07 | | Last modified: | 2018-10-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde
Cell(Cambridge,Mass.), 111, 2002
|
|
1NBH
 
 | |
1NBI
 
 | |
1NBJ
 
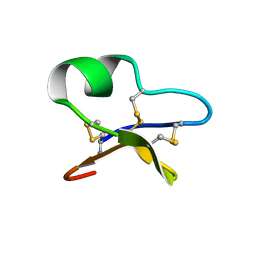 | | High-resolution solution structure of cycloviolacin O1 | | Descriptor: | cycloviolacin O1 | | Authors: | Rosengren, K.J, Daly, N.L, Plan, M.R, Waine, C, Craik, D.J. | | Deposit date: | 2002-12-02 | | Release date: | 2003-03-18 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Twists, Knots, and Rings in Proteins. STRUCTURAL DEFINITION OF THE CYCLOTIDE FRAMEWORK.
J.Biol.Chem., 278, 2003
|
|
1NBK
 
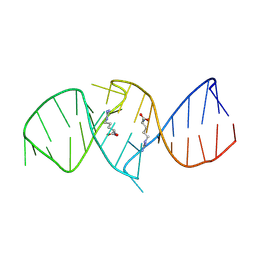 | | The structure of RNA aptamer for HIV Tat complexed with two argininamide molecules | | Descriptor: | 2-AMINO-5-GUANIDINO-PENTANOIC ACID, RNA aptamer | | Authors: | Matsugami, A, Kobayashi, S, Ouhashi, K, Uesugi, S, Yamamoto, R, Taira, K, Nishikawa, S, Kumar, P.K.R, Katahira, M. | | Deposit date: | 2002-12-03 | | Release date: | 2003-12-03 | | Last modified: | 2024-09-18 | | Method: | SOLUTION NMR | | Cite: | Structural Basis of the Highly Efficient Trapping of the HIV Tat Protein by an RNA Aptamer
Structure, 11, 2003
|
|
1NBL
 
 | |
1NBM
 
 | | THE STRUCTURE OF BOVINE F1-ATPASE COVALENTLY INHIBITED WITH 4-CHLORO-7-NITROBENZOFURAZAN | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, F1-ATPASE, ... | | Authors: | Orriss, G.L, Leslie, A.G.W, Braig, K, Walker, J.E. | | Deposit date: | 1998-04-30 | | Release date: | 1998-08-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Bovine F1-ATPase covalently inhibited with 4-chloro-7-nitrobenzofurazan: the structure provides further support for a rotary catalytic mechanism.
Structure, 6, 1998
|
|
1NBO
 
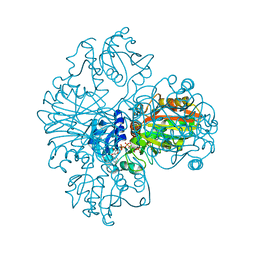 | | The dual coenzyme specificity of photosynthetic glyceraldehyde-3-phosphate dehydrogenase interpreted by the crystal structure of A4 isoform complexed with NAD | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, glyceraldehyde-3-phosphate dehydrogenase A | | Authors: | Falini, G, Fermani, S, Ripamonti, A, Sabatino, P, Sparla, F, Pupillo, P, Trost, P. | | Deposit date: | 2002-12-03 | | Release date: | 2003-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dual Coenzyme Specificity of Photosynthetic Glyceraldehyde-3-phosphate
Dehydrogenase Interpreted by the Crystal Structure of A(4) Isoform
Complexed with NAD
Biochemistry, 42, 2003
|
|
1NBP
 
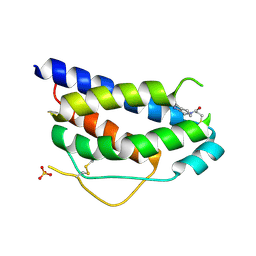 | | Crystal Structure Of Human Interleukin-2 Y31C Covalently Modified At C31 With 3-Mercapto-1-(1,3,4,9-tetrahydro-B-carbolin-2-yl)-propan-1-one | | Descriptor: | 3-MERCAPTO-1-(1,3,4,9-TETRAHYDRO-B-CARBOLIN-2-YL)-PROPAN-1-ONE, Interleukin-2, SULFATE ION | | Authors: | Hyde, J, Braisted, A.C, Randal, M, Arkin, M.R. | | Deposit date: | 2002-12-03 | | Release date: | 2002-12-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery and characterization of cooperative ligand binding in the adaptive region of interleukin-2
Biochemistry, 42, 2003
|
|
1NBQ
 
 | | Crystal Structure of Human Junctional Adhesion Molecule Type 1 | | Descriptor: | Junctional adhesion molecule 1 | | Authors: | Prota, A.E, Campbell, J.A, Schelling, P, Forrest, J.C, Watson, M.J, Peters, T.R, Aurrand-Lions, M, Imhof, B.A, Dermody, T.S, Stehle, T. | | Deposit date: | 2002-12-03 | | Release date: | 2003-04-01 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human junctional adhesion molecule 1: Implications for reovirus binding
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1NBR
 
 | |
