7X38
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 8A10 (CVB1-E:8A10) | | Descriptor: | 8A10 heavy chain, 8A10 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
4DLV
 
 | | H-Ras Set 2 CaCl2/DTT, ordered off | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, CALCIUM ION, GTPase HRas, ... | | Authors: | Holzapfel, G, Mattos, C. | | Deposit date: | 2012-02-06 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.572 Å) | | Cite: | Shift in the Equilibrium between On and Off States of the Allosteric Switch in Ras-GppNHp Affected by Small Molecules and Bulk Solvent Composition.
Biochemistry, 51, 2012
|
|
4J68
 
 | |
7X3F
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 9A3 (CVB1-A:9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.52 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
4J6C
 
 | |
7X2G
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb nAb 2E6 (CVB1-E:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-25 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
4J9I
 
 | |
4JA3
 
 | | Partially occluded inward open conformation of the xylose transporter XylE from E. coli | | Descriptor: | CADMIUM ION, D-xylose-proton symporter, LUTETIUM (III) ION | | Authors: | Quistgaard, E.M, Low, C, Moberg, P, Tresaugues, L, Nordlund, P. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Structural basis for substrate transport in the GLUT-homology family of monosaccharide transporters.
Nat.Struct.Mol.Biol., 20, 2013
|
|
4B1E
 
 | | New Aminoimidazoles as BACE-1 Inhibitors: From Rational Design to Ab- lowering in Brain | | Descriptor: | (2R)-2-methyl-5-phenyl-2-(3-pyridin-3-ylphenyl)-2,3-dihydro-1H-imidazol-4-amine, BETA-SECRETASE 1 | | Authors: | Rahm, F, Blid, J, Ginman, T, Karlstrom, S, Kihlstrom, J, Kolmodin, K, Lindstrom, J, von Berg, S, von Kieseritzky, F, Slivo, C, Swahn, B, Viklund, J, Olsson, L, Johansson, P, Eketjall, S, Falting, J, Jeppsson, F, Stromberg, K, Janson, J, Gravenfors, Y. | | Deposit date: | 2012-07-10 | | Release date: | 2012-10-10 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New aminoimidazoles as beta-secretase (BACE-1) inhibitors showing amyloid-beta (A beta ) lowering in brain.
J. Med. Chem., 55, 2012
|
|
2X9P
 
 | | X-ray structure of the substrate-free cytochrome P450 PimD - a polyene macrolide antibiotic pimaricin epoxidase | | Descriptor: | PIMD PROTEIN, PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION | | Authors: | Kells, P.M, Ouellet, H, Santos-Aberturas, J, Aparicio, J.F, Podust, L.M. | | Deposit date: | 2010-03-23 | | Release date: | 2010-08-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of Cytochrome P450 Pimd Suggests Epoxidation of the Polyene Macrolide Pimaricin Occurs Via a Hydroperoxoferric Intermediate.
Chem.Biol., 17, 2010
|
|
4JA4
 
 | | Inward open conformation of the xylose transporter XylE from E. coli | | Descriptor: | CADMIUM ION, D-xylose-proton symporter | | Authors: | Quistgaard, E.M, Low, C, Moberg, P, Tresaugues, L, Nordlund, P. | | Deposit date: | 2013-02-18 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | Structural basis for substrate transport in the GLUT-homology family of monosaccharide transporters.
Nat.Struct.Mol.Biol., 20, 2013
|
|
3PYH
 
 | | Bovine trypsin variant X(tripleGlu217Ile227) in complex with small molecule inhibitor | | Descriptor: | 3-(3-carbamimidoylphenyl)-N-(2'-sulfamoylbiphenyl-4-yl)-1,2-oxazole-4-carboxamide, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tziridis, A, Neumann, P, Kolenko, P, Stubbs, M.T. | | Deposit date: | 2010-12-13 | | Release date: | 2011-12-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Correlating structure and ligand affinity in drug discovery: a cautionary tale involving second shell residues.
Biol.Chem., 395, 2014
|
|
4DUD
 
 | | cytochrome P450 BM3h-2G9C6 MRI sensor, no ligand | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 variant 2G9C6 | | Authors: | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
4B2S
 
 | | Solution structure of CCP modules 11-12 of complement factor H | | Descriptor: | COMPLEMENT FACTOR H | | Authors: | Makou, E, Mertens, H.D, Maciejewski, M, Soares, D.C, Matis, I, Schmidt, C.Q, Herbert, A.P, Svergun, D.I, Barlow, P.N. | | Deposit date: | 2012-07-17 | | Release date: | 2012-10-17 | | Last modified: | 2013-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Ccp Modules 10-12 Illuminates Functional Architecture of the Complement Regulator, Factor H.
J.Mol.Biol., 424, 2012
|
|
4BAD
 
 | | Hen egg-white lysozyme structure in complex with the europium tris- hydroxymethyltriazoledipicolinate complex at 1.35 A resolution. | | Descriptor: | 4-(4-(hydroxymethyl)-1h-1,2,3-triazol-1-yl)pyridine-2,6-dicarboxylic acid, ACETATE ION, CHLORIDE ION, ... | | Authors: | Talon, R, Kahn, R, Gautier, A, Nauton, L, Girard, E. | | Deposit date: | 2012-09-13 | | Release date: | 2012-11-14 | | Last modified: | 2019-03-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Clicked Europium Dipicolinate Complexes for Protein X-Ray Structure Determination.
Chem.Commun.(Camb.), 48, 2012
|
|
4DUC
 
 | | cytochrome P450 BM3h-2G9 MRI sensor, no ligand | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 BM3 variant 2G9 | | Authors: | Brustad, E.M, Lelyveld, V.S, Snow, C.D, Crook, N, Martinez, F.M, Scholl, T.J, Jasanoff, A, Arnold, F.H. | | Deposit date: | 2012-02-21 | | Release date: | 2012-06-13 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure-guided directed evolution of highly selective p450-based magnetic resonance imaging sensors for dopamine and serotonin.
J.Mol.Biol., 422, 2012
|
|
7X37
 
 | | Cryo-EM structure of Coxsackievirus B1 A particle in complex with nAb 2E6 (CVB1-A:2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-02-28 | | Release date: | 2022-09-28 | | Last modified: | 2022-10-05 | | Method: | ELECTRON MICROSCOPY (3.31 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
4DX1
 
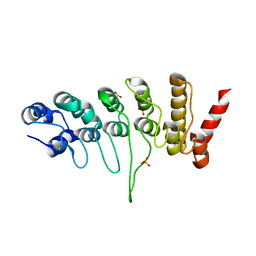 | | Crystal structure of the human TRPV4 ankyrin repeat domain | | Descriptor: | GLYCEROL, PHOSPHATE ION, Transient receptor potential cation channel subfamily V member 4 | | Authors: | Inada, H, Gaudet, R. | | Deposit date: | 2012-02-27 | | Release date: | 2012-07-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural and biochemical consequences of disease-causing mutations in the ankyrin repeat domain of the human TRPV4 channel.
Biochemistry, 51, 2012
|
|
4B7N
 
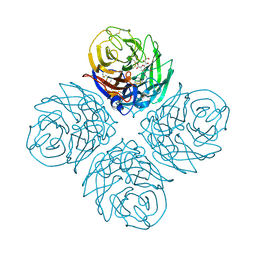 | | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, NEURAMINIDASE, ... | | Authors: | van der Vries, E, Vachieri, S.G, Xiong, X, Liu, J, Collins, P.J, Walker, P.A, Haire, L.F, Hay, A.J, Schutten, M, Osterhaus, A.D.M.E, Martin, S.R, Boucher, C.A.B, Skehel, J.J, Gamblin, S.J. | | Deposit date: | 2012-08-21 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | H1N1 2009 Pandemic Influenza Virus: Resistance of the I223R Neuraminidase Mutant Explained by Kinetic and Structural Analysis
Plos Pathog., 8, 2012
|
|
2WQ5
 
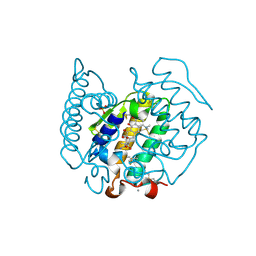 | | Non-antibiotic properties of tetracyclines: structural basis for inhibition of secretory phospholipase A2. | | Descriptor: | (4S,4AS,5AR,12AS)-4,7-BIS(DIMETHYLAMINO)-3,10,12,12A-TETRAHYDROXY-1,11-DIOXO-1,4,4A,5,5A,6,11,12A-OCTAHYDROTETRACENE-2- CARBOXAMIDE, CALCIUM ION, PHOSPHOLIPASE A2, ... | | Authors: | Dalm, D, Palm, G.J, Hinrichs, W. | | Deposit date: | 2009-08-13 | | Release date: | 2010-03-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Non-Antibiotic Properties of Tetracyclines: Structural Basis for Inhibition of Secretory Phospholipase A(2).
J.Mol.Biol., 398, 2010
|
|
4DW2
 
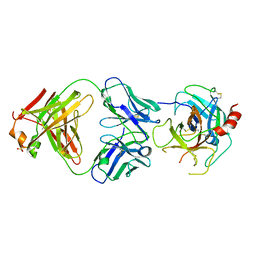 | | The crystal structure of uPA in complex with the Fab fragment of mAb-112 | | Descriptor: | Fab fragment of pro-uPA antibody mAb-112, SULFATE ION, Urokinase-type plasminogen activator | | Authors: | Jiang, L, Botkjaer, K.A, Andersen, L.M, Yuan, C, Andreasen, P.A, Huang, M. | | Deposit date: | 2012-02-24 | | Release date: | 2013-01-16 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Rezymogenation of active urokinase induced by an inhibitory antibody.
Biochem.J., 449, 2013
|
|
7X46
 
 | | Cryo-EM structure of Coxsackievirus B1 A-particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, VP2, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.85 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X47
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 2E6 (classified from CVB1 mature virion in complex with 8A10 and 2E6) | | Descriptor: | 2E6 heavy chain, 2E6 light chain, Genome polyprotein, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.66 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
7X4K
 
 | | Cryo-EM structure of Coxsackievirus B1 empty particle in complex with nAb 9A3 (classified from CVB1 mature virion in complex with 8A10 and 9A3) | | Descriptor: | 9A3 heavy chain, 9A3 light chain, Genome polyprotein, ... | | Authors: | Zheng, Q, Zhu, R, Sun, H, Cheng, T, Li, S, Xia, N. | | Deposit date: | 2022-03-02 | | Release date: | 2022-09-28 | | Method: | ELECTRON MICROSCOPY (3.82 Å) | | Cite: | Structural basis for the synergistic neutralization of coxsackievirus B1 by a triple-antibody cocktail.
Cell Host Microbe, 30, 2022
|
|
3Q2L
 
 | | Mouse E-cadherin EC1-2 V81D mutant | | Descriptor: | CALCIUM ION, Cadherin-1, PENTAETHYLENE GLYCOL | | Authors: | Harrison, O.J, Jin, X, Shapiro, L. | | Deposit date: | 2010-12-20 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The extracellular architecture of adherens junctions revealed by crystal structures of type I cadherins.
Structure, 19, 2011
|
|
