2Y5K
 
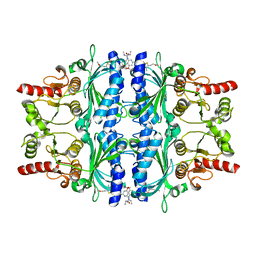 | | Orally active aminopyridines as inhibitors of tetrameric fructose 1,6- bisphosphatase | | Descriptor: | 1-[5-(2-METHOXYETHYL)-4-METHYL-THIOPHEN-2-YL]SULFONYL-3-[4-METHOXY-6-(METHYLCARBAMOYLAMINO)PYRIDIN-2-YL]UREA, FRUCTOSE-1,6-BISPHOSPHATASE 1 | | Authors: | Ruf, A, Hebeisen, P, Haap, W, Kuhn, B, Mohr, P, Wessel, H.P, Zutter, U, Kirchner, S, Benz, J, Joseph, C, Alvarez-Sanchez, R, Gubler, M, Schott, B, Benardeau, A, Tozzo, E, Kitas, E. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Orally Active Aminopyridines as Inhibitors of Tetrameric Fructose-1,6-Bisphosphatase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
2Y5L
 
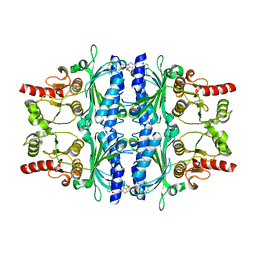 | | orally active aminopyridines as inhibitors of tetrameric fructose 1,6- bisphosphatase | | Descriptor: | FRUCTOSE-1,6-BISPHOSPHATASE 1, N-{[(2Z)-5-bromo-1,3-thiazol-2(3H)-ylidene]carbamoyl}-3-chlorobenzenesulfonamide | | Authors: | ruf, a, hebeisen, p, haap, w, kuhn, b, mohr, p, wessel, h.p, zutter, u, kirchner, s, benz, j, joseph, c, alvarez-sanchez, r, gubler, m, schott, b, benardeau, a, tozzo, e, kitas, e. | | Deposit date: | 2011-01-14 | | Release date: | 2011-05-18 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Orally Active Aminopyridines as Inhibitors of Tetrameric Fructose-1,6-Bisphosphatase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4OZN
 
 | | GlnK2 from Haloferax mediterranei complexed with ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Nitrogen regulatory protein P-II, SULFATE ION | | Authors: | Palanca, C, Pedro-Roig, L, Llacer, J.L, Camacho, M, Bonete, M.J, Rubio, V. | | Deposit date: | 2014-02-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The structure of a PII signaling protein from a halophilic archaeon reveals novel traits and high-salt adaptations.
Febs J., 281, 2014
|
|
4OZJ
 
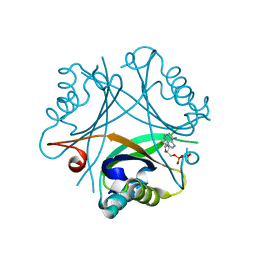 | | GlnK2 from Haloferax mediterranei complexed with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Nitrogen regulatory protein P-II | | Authors: | Palanca, C, Pedro-Roig, L, Llacer, J.L, Camacho, M, Bonete, M.J, Rubio, V. | | Deposit date: | 2014-02-17 | | Release date: | 2014-07-02 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | The structure of a PII signaling protein from a halophilic archaeon reveals novel traits and high-salt adaptations.
Febs J., 281, 2014
|
|
3U16
 
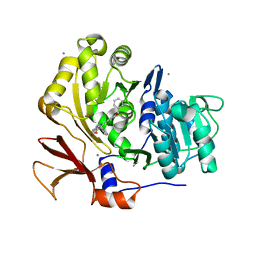 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-(p-benzyloxy)phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-[4-(benzyloxy)phenyl]-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | Authors: | Gulick, A.M, Drake, E.J, Aldrich, C.C, Neres, J. | | Deposit date: | 2011-09-29 | | Release date: | 2012-10-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Non-nucleoside inhibitors of BasE, an adenylating enzyme in the siderophore biosynthetic pathway of the opportunistic pathogen Acinetobacter baumannii.
J.Med.Chem., 56, 2013
|
|
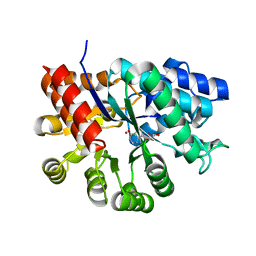
3KM8
 
 | |
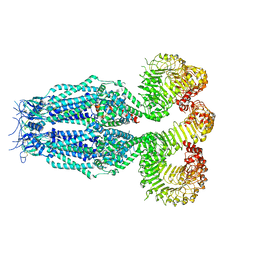
8B42
 
 | |
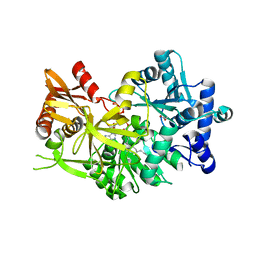
7R7G
 
 | |
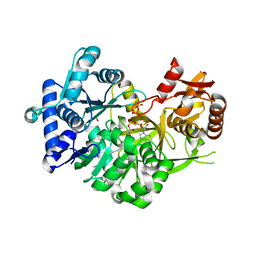
7R7F
 
 | |
7R2Y
 
 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803 in complex with ATP resulting from short ATP soak, conflicting T-loop and C-loop with partial occupancy | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Membrane-associated protein slr1513, SODIUM ION | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7R2Z
 
 | |
7R32
 
 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803, delta104 variant, in complex with ADP (co-crystal), tetragonal crystal form | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Membrane-associated protein slr1513 | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7R31
 
 | | Carbon regulatory PII-like protein SbtB from Synechocystis sp. 6803, C105A+C110A variant, in complex with ATP (co-crystal), tetragonal crystal form | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, Membrane-associated protein slr1513, ... | | Authors: | Selim, K.A, Albrecht, R, Hartmann, M.D. | | Deposit date: | 2022-02-06 | | Release date: | 2023-02-15 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Carbon signaling protein SbtB possesses atypical redox-regulated apyrase activity to facilitate regulation of bicarbonate transporter SbtA.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6DJB
 
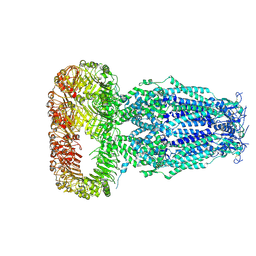 | | Structure of human Volume Regulated Anion Channel composed of SWELL1 (LRRC8A) | | Descriptor: | Volume-regulated anion channel subunit LRRC8A | | Authors: | Kefauver, J.M, Saotome, K, Pallesen, J, Cottrell, C.A, Ward, A.B, Patapoutian, A. | | Deposit date: | 2018-05-24 | | Release date: | 2018-08-15 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of the human volume regulated anion channel.
Elife, 7, 2018
|
|
8HIJ
 
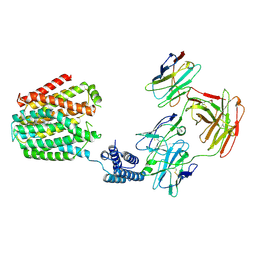 | | The 5-MTHF-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HIK
 
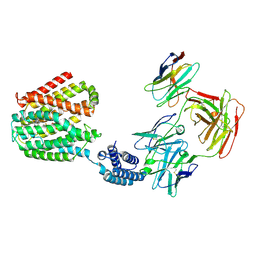 | | The TPP-bound BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | Anti-BRIL Fab heavy chain, Anti-BRIL Fab light chain, Anti-Fab nanobody, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.72 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
8HII
 
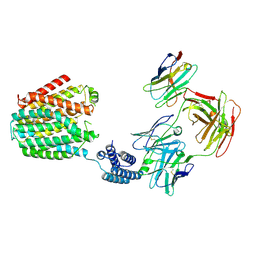 | | The BRIL-SLC19A1/Fab/Nb ternary complex | | Descriptor: | BRIL-SLC19A1 chimera, anti-BRIL Fab heavy chain, anti-BRIL Fab light chain, ... | | Authors: | Zhang, Z, Dang, Y. | | Deposit date: | 2022-11-20 | | Release date: | 2022-12-21 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | Molecular mechanism of substrate recognition by folate transporter SLC19A1.
Cell Discov, 8, 2022
|
|
3LUK
 
 | |
6KX3
 
 | | Crystal structure of RhoA protein with covalent inhibitor DC-Rhoin | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA, prop-2-enyl (3R)-1,1-bis(oxidanylidene)-2,3-dihydro-1-benzothiophene-3-carboxylate | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.981 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
6KX2
 
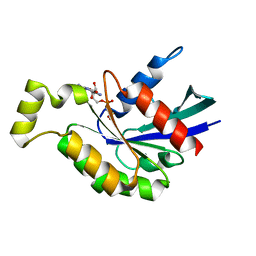 | | Crystal structure of GDP bound RhoA protein | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Transforming protein RhoA | | Authors: | Zhang, H, Luo, C. | | Deposit date: | 2019-09-09 | | Release date: | 2020-08-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Covalent Inhibitors Allosterically Block the Activation of Rho Family Proteins and Suppress Cancer Cell Invasion.
Adv Sci, 7, 2020
|
|
7RTG
 
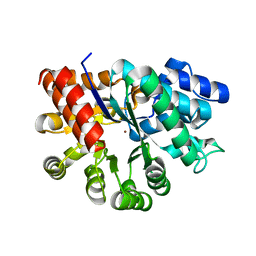 | | Crystal Structure of the Human Adenosine Deaminase 1 | | Descriptor: | Adenosine deaminase, ZINC ION | | Authors: | Ma, M.T, Lieberman, R.L, Blazeck, J, Jennings, M.R. | | Deposit date: | 2021-08-13 | | Release date: | 2022-01-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.591 Å) | | Cite: | Catalytically active holo Homo sapiens adenosine deaminase I adopts a closed conformation.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
5DV9
 
 | |
4FL3
 
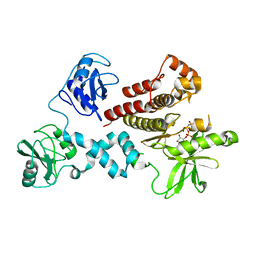 | | Structural and Biophysical Characterization of the Syk Activation Switch | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tyrosine-protein kinase SYK | | Authors: | Graedler, U, Schwarz, D, Dresing, V, Musil, M, Bomke, J, Frech, M, Jaekel, S, Rysiok, T, Mueller-Pompalla, D, Wegener, A. | | Deposit date: | 2012-06-14 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and biophysical characterization of the syk activation switch.
J.Mol.Biol., 425, 2013
|
|
4FL1
 
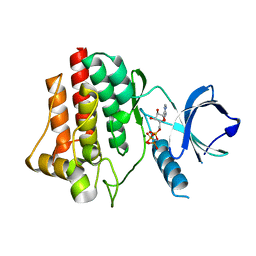 | | Structural and Biophysical Characterization of the Syk Activation Switch | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tyrosine-protein kinase SYK | | Authors: | Graedler, U, Schwarz, D, Dresing, V, Musil, M, Bomke, J, Frech, M, Jaekel, S, Rysiok, T, Mueller-Pompalla, D, Wegener, A. | | Deposit date: | 2012-06-14 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Structural and biophysical characterization of the syk activation switch.
J.Mol.Biol., 425, 2013
|
|
8SV6
 
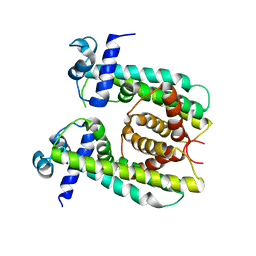 | | Structure of the M. smegmatis DarR protein | | Descriptor: | Fatty acid metabolism regulator protein | | Authors: | Schumacher, M.A. | | Deposit date: | 2023-05-15 | | Release date: | 2023-11-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Structures of the DarR transcription regulator reveal unique modes of second messenger and DNA binding.
Nat Commun, 14, 2023
|
|
