7OBK
 
 | |
7OBL
 
 | |
7OBC
 
 | |
1RK5
 
 | | The D-aminoacylase mutant D366A in complex with 100mM CuCl2 | | Descriptor: | ACETATE ION, COPPER (II) ION, D-aminoacylase, ... | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
4IS3
 
 | |
3T3S
 
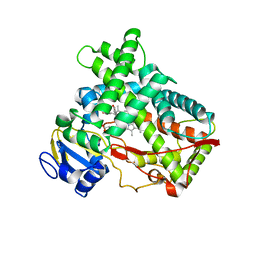 | | Human Cytochrome P450 2A13 in complex with Pilocarpine | | Descriptor: | (3S,4R)-3-ethyl-4-[(1-methyl-1H-imidazol-5-yl)methyl]dihydrofuran-2(3H)-one, Cytochrome P450 2A13, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | DeVore, N.M, Scott, E.E. | | Deposit date: | 2011-07-25 | | Release date: | 2011-12-07 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural comparison of cytochromes P450 2A6, 2A13, and 2E1 with pilocarpine.
Febs J., 279, 2012
|
|
5B7M
 
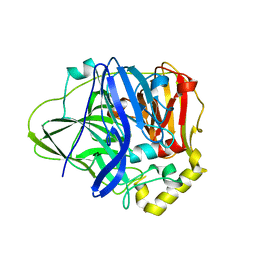 | | Structure of perdeuterated CueO - the signal peptide was truncated by HRV3C protease | | Descriptor: | Blue copper oxidase CueO, COPPER (II) ION | | Authors: | Akter, M, Higuchi, Y, Shibata, N. | | Deposit date: | 2016-06-07 | | Release date: | 2016-10-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biochemical, spectroscopic and X-ray structural analysis of deuterated multicopper oxidase CueO prepared from a new expression construct for neutron crystallography
Acta Crystallogr.,Sect.F, 72, 2016
|
|
3SQ5
 
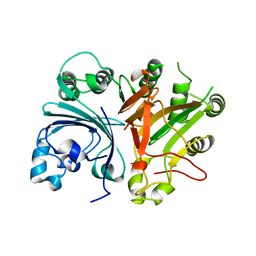 | |
3OJC
 
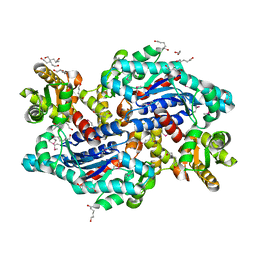 | | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis | | Descriptor: | CALCIUM ION, HEXANE-1,6-DIOL, Putative aspartate/glutamate racemase | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Anderson, W.F, Savchenko, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-08-21 | | Release date: | 2010-09-15 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative Asp/Glu Racemase from Yersinia pestis
To be Published
|
|
3P0R
 
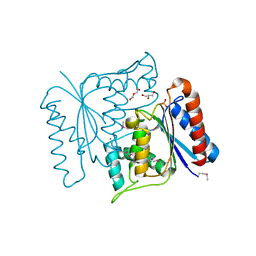 | | Crystal structure of azoreductase from Bacillus anthracis str. Sterne | | Descriptor: | Azoreductase, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Filippova, E.V, Wawrzak, Z, Kudritska, M, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-09-29 | | Release date: | 2010-10-20 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Crystal structure of azoreductase from Bacillus anthracis str. Sterne
To be Published
|
|
4I3Q
 
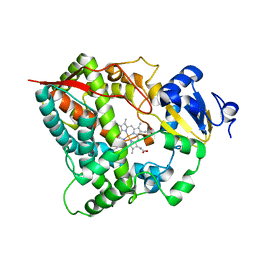 | |
4I91
 
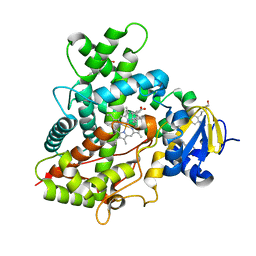 | | Crystal Structure of Cytochrome P450 2B6 (Y226H/K262R) in complex with alpha-Pinene. | | Descriptor: | (+)-alpha-Pinene, 5-CYCLOHEXYL-1-PENTYL-BETA-D-MALTOSIDE, Cytochrome P450 2B6, ... | | Authors: | Shah, M.B, Stout, C.D, Halpert, J.R. | | Deposit date: | 2012-12-04 | | Release date: | 2013-07-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Thermodynamic Basis of (+)-alpha-Pinene Binding to Human Cytochrome P450 2B6.
J.Am.Chem.Soc., 135, 2013
|
|
3NXU
 
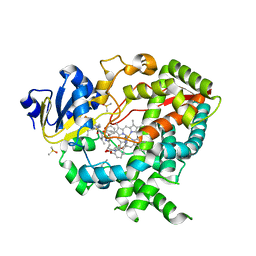 | |
1V51
 
 | | The functional role of the binuclear metal center in D-aminoacylase. One-metal activation and second-metal attenuation | | Descriptor: | ACETATE ION, D-aminoacylase, ZINC ION | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The functional role of the binuclear metal center in D-aminoacylase: one-metal activation and second-metal attenuation.
J.Biol.Chem., 279, 2004
|
|
1UP7
 
 | | Structure of the 6-phospho-beta glucosidase from Thermotoga maritima at 2.4 Angstrom resolution in the tetragonal form with NAD and glucose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-glucopyranose, 6-PHOSPHO-BETA-GLUCOSIDASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Varrot, A, Yip, V.L, Withers, S.G, Davies, G.J. | | Deposit date: | 2003-09-29 | | Release date: | 2004-11-18 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Nad+ and Metal-Ion Dependent Hydrolysis by Family 4 Glycosidases: Structural Insight Into Specificity for Phospho-Beta-D-Glucosides
J.Mol.Biol., 346, 2005
|
|
2WFW
 
 | | Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases - The Arc domain structure | | Descriptor: | ARC | | Authors: | Djuranovic, S, Hartmann, M.D, Habeck, M, Ursinus, A, Zwickl, P, Martin, J, Lupas, A.N, Zeth, K. | | Deposit date: | 2009-04-15 | | Release date: | 2009-05-12 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
3PMS
 
 | |
2WG5
 
 | | Proteasome-Activating Nucleotidase (PAN) N-domain (57-134) from Archaeoglobus fulgidus fused to GCN4 | | Descriptor: | GENERAL CONTROL PROTEIN GCN4, PROTEASOME-ACTIVATING NUCLEOTIDASE | | Authors: | Hartmann, M.D, Djuranovic, S, Ursinus, A, Zeth, K, Lupas, A.N. | | Deposit date: | 2009-04-15 | | Release date: | 2009-04-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Activity of the N-Terminal Substrate Recognition Domains in Proteasomal Atpases.
Mol.Cell, 34, 2009
|
|
3MVR
 
 | |
3SQ3
 
 | |
3TBG
 
 | | Human cytochrome P450 2D6 with two thioridazines bound in active site | | Descriptor: | 10-{2-[(2R)-1-methylpiperidin-2-yl]ethyl}-2-(methylsulfanyl)-10H-phenothiazine, Cytochrome P450 2D6, GLYCEROL, ... | | Authors: | Wang, A, Stout, C.D, Johnson, E.F. | | Deposit date: | 2011-08-05 | | Release date: | 2012-08-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Contributions of Ionic Interactions and Protein Dynamics to Cytochrome P450 2D6 (CYP2D6) Substrate and Inhibitor Binding.
J.Biol.Chem., 290, 2015
|
|
1B73
 
 | | GLUTAMATE RACEMASE FROM AQUIFEX PYROPHILUS | | Descriptor: | GLUTAMATE RACEMASE | | Authors: | Hwang, K.Y, Cho, C.S, Kim, S.S, Yu, Y.G, Cho, Y. | | Deposit date: | 1999-01-26 | | Release date: | 1999-01-28 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and mechanism of glutamate racemase from Aquifex pyrophilus.
Nat.Struct.Biol., 6, 1999
|
|
1TFR
 
 | | RNASE H FROM BACTERIOPHAGE T4 | | Descriptor: | MAGNESIUM ION, T4 RNASE H | | Authors: | Mueser, T.C, Nossal, N.G, Hyde, C.C. | | Deposit date: | 1996-04-27 | | Release date: | 1996-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure of bacteriophage T4 RNase H, a 5' to 3' RNA-DNA and DNA-DNA exonuclease with sequence similarity to the RAD2 family of eukaryotic proteins.
Cell(Cambridge,Mass.), 85, 1996
|
|
1TQN
 
 | | Crystal Structure of Human Microsomal P450 3A4 | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, cytochrome P450 3A4 | | Authors: | Yano, J.K, Wester, M.R, Schoch, G.A, Griffin, K.J, Stout, C.D, Johnson, E.F. | | Deposit date: | 2004-06-17 | | Release date: | 2004-07-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Structure of Human Microsomal Cytochrome P450 3A4 Determined by X-ray Crystallography to 2.05-A Resolution
J.Biol.Chem., 279, 2004
|
|
1V4Y
 
 | | The functional role of the binuclear metal center in D-aminoacylase. One-metal activation and second-metal attenuation | | Descriptor: | ACETATE ION, D-aminoacylase, ZINC ION | | Authors: | Lai, W.L, Chou, L.Y, Ting, C.Y, Tsai, Y.C, Liaw, S.H. | | Deposit date: | 2003-11-20 | | Release date: | 2004-04-20 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Functional Role of the Binuclear Metal Center in D-Aminoacylase: ONE-METAL ACTIVATION AND SECOND-METAL ATTENUATION.
J.Biol.Chem., 279, 2004
|
|
