3SGW
 
 | |
5LTZ
 
 | | GmhA_mutant Q175E | | Descriptor: | 1,2-ETHANEDIOL, D-ALTRO-HEPT-2-ULOSE 7-PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Vivoli, M, Harmer, N.J. | | Deposit date: | 2016-09-07 | | Release date: | 2017-12-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A half-site multimeric enzyme achieves its cooperativity without conformational changes.
Sci Rep, 7, 2017
|
|
1O8J
 
 | |
1O8G
 
 | |
3SKF
 
 | |
8DOV
 
 | | Crystal structure of the Shr Hemoglobin Interacting Domain 2 (HID2) in complex with Hemoglobin | | Descriptor: | GLYCEROL, Heme-binding protein Shr, Hemoglobin subunit alpha, ... | | Authors: | Macdonald, R, Mahoney, B.J, Cascio, D, Clubb, R.T. | | Deposit date: | 2022-07-14 | | Release date: | 2023-01-25 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The Shr receptor from Streptococcus pyogenes uses a cap and release mechanism to acquire heme-iron from human hemoglobin.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1CHI
 
 | |
6H2E
 
 | |
5MTR
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT512 | | Descriptor: | 2-[4-[(4-cyclopentyl-1,2,3-triazol-1-yl)methyl]-2-oxidanyl-phenoxy]benzenecarbonitrile, CHLORIDE ION, Enoyl-[acyl-carrier-protein] reductase [NADH], ... | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
6H7E
 
 | | GEF regulatory domain | | Descriptor: | ADENOSINE-3',5'-CYCLIC-MONOPHOSPHATE, SULFATE ION, cDNA FLJ56134, ... | | Authors: | Ferrandez, Y, Cherfils, J, Peurois, F. | | Deposit date: | 2018-07-31 | | Release date: | 2020-02-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Membranes prime the RapGEF EPAC1 to transduce cAMP signaling.
Nat Commun, 14, 2023
|
|
1OFK
 
 | |
8CJC
 
 | | F515A variant of the CODH/ACS complex of C. hydrogenoformans | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, CO-methylating acetyl-CoA synthase, ... | | Authors: | Ruickoldt, J, Jeoung, J, Lennartz, F, Dobbek, H. | | Deposit date: | 2023-02-13 | | Release date: | 2024-02-21 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Coupling CO 2 Reduction and Acetyl-CoA Formation: The Role of a CO Capturing Tunnel in Enzymatic Catalysis.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
1ON7
 
 | |
5MTQ
 
 | | Crystal structure of M. tuberculosis InhA inhibited by PT511 | | Descriptor: | 2-[4-[(4-cyclohexyl-1,2,3-triazol-1-yl)methyl]-2-oxidanyl-phenoxy]benzenecarbonitrile, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Eltschkner, S, Pschibul, A, Spagnuolo, L.A, Yu, W, Tonge, P.J, Kisker, C. | | Deposit date: | 2017-01-10 | | Release date: | 2017-02-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Evaluating the Contribution of Transition-State Destabilization to Changes in the Residence Time of Triazole-Based InhA Inhibitors.
J. Am. Chem. Soc., 139, 2017
|
|
5M6B
 
 | |
6XDK
 
 | |
3SNY
 
 | |
3SMR
 
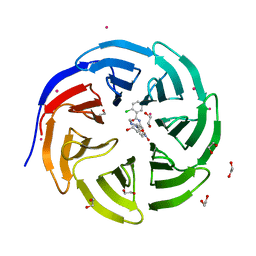 | | Crystal structure of human WD repeat domain 5 with compound | | Descriptor: | 1,2-ETHANEDIOL, 2-chloro-N-[2-(4-methylpiperazin-1-yl)-5-nitrophenyl]benzamide, UNKNOWN ATOM OR ION, ... | | Authors: | Dong, A, Dombrovski, L, Wasney, G.A, Tempel, W, Senisterra, G, Bolshan, Y, Smil, D, Nguyen, K.T, Hajian, T, Poda, G, Al-Awar, R, Bountra, C, Weigelt, J, Edwards, A.M, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Small-molecule inhibition of MLL activity by disruption of its interaction with WDR5.
Biochem. J., 449, 2013
|
|
3SO1
 
 | |
5LMH
 
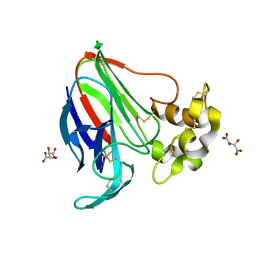 | | High dose Thaumatin - 160-200 ms. | | Descriptor: | L(+)-TARTARIC ACID, Thaumatin-1 | | Authors: | Schubert, R, Kapis, S, Heymann, M, Giquel, Y, Bourenkov, G, Schneider, T, Betzel, C, Perbandt, M. | | Deposit date: | 2016-07-30 | | Release date: | 2016-11-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | A multicrystal diffraction data-collection approach for studying structural dynamics with millisecond temporal resolution.
IUCrJ, 3, 2016
|
|
6HMM
 
 | | POLYADPRIBOSYL GLYCOHYDROLASE IN COMPLEX WITH PDD00013907 | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, Poly(ADP-ribose) glycohydrolase, ... | | Authors: | Tucker, J.A, Brassington, C, Hassall, G. | | Deposit date: | 2018-09-12 | | Release date: | 2018-11-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Cell-Active Small Molecule Inhibitors of the DNA-Damage Repair Enzyme Poly(ADP-ribose) Glycohydrolase (PARG): Discovery and Optimization of Orally Bioavailable Quinazolinedione Sulfonamides.
J.Med.Chem., 61, 2018
|
|
6HSQ
 
 | |
1TVK
 
 | | The binding mode of epothilone A on a,b-tubulin by electron crystallography | | Descriptor: | EPOTHILONE A, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Nettles, J.H, Li, H, Cornett, B, Krahn, J.M, Snyder, J.P, Downing, K.H. | | Deposit date: | 2004-06-29 | | Release date: | 2004-09-14 | | Last modified: | 2023-08-23 | | Method: | ELECTRON CRYSTALLOGRAPHY (2.89 Å) | | Cite: | The binding mode of epothilone A on alpha,beta-tubulin by electron crystallography
Science, 305, 2004
|
|
6HSA
 
 | |
1RMS
 
 | | CRYSTAL STRUCTURES OF RIBONUCLEASE MS COMPLEXED WITH 3'-GUANYLIC ACID A GP*C ANALOGUE, 2'-DEOXY-2'-FLUOROGUANYLYL-3',5'-CYTIDINE | | Descriptor: | GUANOSINE-3'-MONOPHOSPHATE, RIBONUCLEASE MS | | Authors: | Nonaka, T, Mitsui, Y, Nakamura, K.T. | | Deposit date: | 1991-12-02 | | Release date: | 1992-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ribonuclease Ms (as a ribonuclease T1 homologue) complexed with a guanylyl-3',5'-cytidine analogue.
Biochemistry, 32, 1993
|
|
