6E0N
 
 | | Structure of Elizabethkingia meningoseptica CdnE cyclic dinucleotide synthase with GTP and Apcpp | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
6E0O
 
 | | Structure of Elizabethkingia meningoseptica CdnE cyclic dinucleotide synthase with pppA[3'-5']pA | | Descriptor: | MAGNESIUM ION, RNA (5'-D(*(ATP))-R(P*A)-3'), cGAS/DncV-like nucleotidyltransferase in E. coli homolog | | Authors: | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | Deposit date: | 2018-07-06 | | Release date: | 2019-02-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
6E0P
 
 | |
6E0Q
 
 | | The N-terminal domain of PA endonuclease from the influenza H1N1 virus in complex with 4,6-dihydroxy-2-methyl-5-oxocyclohepta-1,3,6-triene-1-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 4,6-dihydroxy-2-methyl-5-oxocyclohepta-1,3,6-triene-1-carboxylic acid, MANGANESE (II) ION, ... | | Authors: | Dick, B.L, Morrison, C.N, Cohen, S.M. | | Deposit date: | 2018-07-06 | | Release date: | 2018-11-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structure-Activity Relationships in Metal-Binding Pharmacophores for Influenza Endonuclease.
J. Med. Chem., 61, 2018
|
|
6E0R
 
 | | hALK in complex with compound 7 N-((1S)-1-(5-fluoropyridin-2-yl)ethyl)-1-(5-methyl-1H-pyrazol-3-yl)-3-(oxetan-3-ylsulfonyl)-1H-pyrrolo[2,3-b]pyridin-6-amine | | Descriptor: | ALK tyrosine kinase receptor, N-[(1S)-1-(5-fluoropyridin-2-yl)ethyl]-1-(5-methyl-1H-pyrazol-3-yl)-3-[(oxetan-3-yl)sulfonyl]-1H-pyrrolo[2,3-b]pyridin-6-amine | | Authors: | Lane, W, Saikatendu, K. | | Deposit date: | 2018-07-06 | | Release date: | 2019-05-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Discovery of Potent, Selective, and Brain-Penetrant 1 H-Pyrazol-5-yl-1 H-pyrrolo[2,3- b]pyridines as Anaplastic Lymphoma Kinase (ALK) Inhibitors.
J.Med.Chem., 62, 2019
|
|
6E0S
 
 | | Crystal structure of MEM-A1, a subclass B3 metallo-beta-lactamase isolated from a soil metagenome library | | Descriptor: | GLYCEROL, MEM-A1, PHOSPHATE ION, ... | | Authors: | Stogios, P.J, Skarina, T, Lau, C, Topp, E, Savchenko, A, Joachimiak, A, Satchell, K.J, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2018-07-06 | | Release date: | 2018-07-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of MEM-A1, a subclass B3 metallo-beta-lactamase isolated from a soil metagenome library
To Be Published
|
|
6E0T
 
 | | C-terminal domain of Fission Yeast OFD1 | | Descriptor: | Prolyl 3,4-dihydroxylase ofd1 | | Authors: | Bianchet, M.A, Amzel, L.M, Espenshade, P.J, Yeh, T. | | Deposit date: | 2018-07-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | The hypoxic regulator of sterol synthesis nro1 is a nuclear import adaptor.
Structure, 19, 2011
|
|
6E0U
 
 | |
6E0V
 
 | | Apo crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 | | Descriptor: | 1,2-ETHANEDIOL, Bacteriophage Phi92 gp150, CHLORIDE ION, ... | | Authors: | Leiman, P.G, Browning, C, Gerardy-Schahn, R, Shneider, M.M, Plattner, M, Schwarzer, D. | | Deposit date: | 2018-07-07 | | Release date: | 2019-07-17 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 complexed with one repeating unit of colanic acid
To Be Published
|
|
6E0W
 
 | | Crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 complexed with one repeating unit of colanic acid | | Descriptor: | 1,2-ETHANEDIOL, 4,6-O-[(1R)-1-carboxyethylidene]-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-alpha-D-galactopyranose-(1-3)-alpha-L-fucopyranose-(1-4)-alpha-L-fucopyranose-(1-3)-beta-D-glucopyranose, Bacteriophage Phi92 gp150, ... | | Authors: | Plattner, M, Browning, C, Gerardy-Schahn, R, Shneider, M.M, Leiman, P.G, Schwarzer, D. | | Deposit date: | 2018-07-07 | | Release date: | 2019-07-17 | | Last modified: | 2025-04-16 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Crystal structure of the colanidase tailspike protein gp150 of Phage Phi92 complexed with one repeating unit of colanic acid
To Be Published
|
|
6E0X
 
 | |
6E0Y
 
 | |
6E0Z
 
 | |
6E10
 
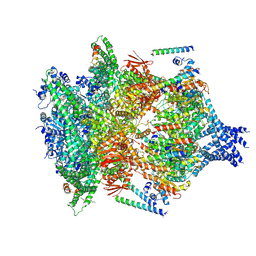 | | PTEX Core Complex in the Engaged (Extended) State | | Descriptor: | Endogenous cargo polypeptide, Exported protein 2, Heat shock protein 101, ... | | Authors: | Ho, C, Lai, M, Zhou, Z.H. | | Deposit date: | 2018-07-08 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.16 Å) | | Cite: | Malaria parasite translocon structure and mechanism of effector export.
Nature, 561, 2018
|
|
6E11
 
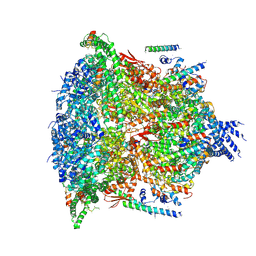 | | PTEX Core Complex in the Resetting (Compact) State | | Descriptor: | Endogenous cargo polypeptide, Exported protein 2, Heat shock protein 101, ... | | Authors: | Ho, C, Lai, M, Zhou, Z.H. | | Deposit date: | 2018-07-08 | | Release date: | 2018-08-22 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Malaria parasite translocon structure and mechanism of effector export.
Nature, 561, 2018
|
|
6E12
 
 | | Crystal Structure of the Alr8543 protein in complex with Oleic Acid and magnesium ion from Nostoc sp. PCC 7120, Northeast Structural Genomics Consortium Target NsR141 | | Descriptor: | Alr8543 protein, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Forouhar, F, Neely, H, Seetharaman, J, Xiao, R, Ciccosanti, C, Patel, D, Belote, R.L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2018-07-09 | | Release date: | 2018-07-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Alr8543 protein in complex with Oleic Acid and magnesium ion from Nostoc sp. PCC 7120, Northeast Structural Genomics Consortium Target NsR141
To Be Published
|
|
6E13
 
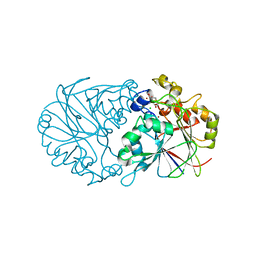 | | Pseudomonas putida PqqB with a non-physiological zinc at the active site binds the substrate mimic, 5-cysteinyl-3,4-dihydroxyphenylalanine (5-Cys-DOPA), non-specifically but supports the proposed function of the enzyme in pyrroloquinoline quinone biosynthesis. | | Descriptor: | 3-{[(2S)-2-amino-2-carboxyethyl]sulfanyl}-5-hydroxy-L-tyrosine, CHLORIDE ION, Coenzyme PQQ synthesis protein B, ... | | Authors: | Evans III, R.L, Wilmot, C.M. | | Deposit date: | 2018-07-09 | | Release date: | 2019-05-22 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | Discovery of Hydroxylase Activity for PqqB Provides a Missing Link in the Pyrroloquinoline Quinone Biosynthetic Pathway.
J.Am.Chem.Soc., 141, 2019
|
|
6E14
 
 | | Handover mechanism of the growing pilus by the bacterial outer membrane usher FimD | | Descriptor: | Chaperone protein FimC, Fimbrial biogenesis outer membrane usher protein, Protein FimF, ... | | Authors: | Du, M, Yuan, Z, Yu, H, Henderson, N, Sarowar, S, Zhao, G, Werneburg, G.T, Thanassi, D.G, Li, H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD.
Nature, 562, 2018
|
|
6E15
 
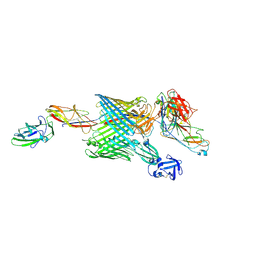 | | Handover mechanism of the growing pilus by the bacterial outer membrane usher FimD | | Descriptor: | Chaperone protein FimC, Fimbrial biogenesis outer membrane usher protein, Protein FimF, ... | | Authors: | Du, M, Yuan, Z, Yu, H, Henderson, N, Sarowar, S, Zhao, G, Werneburg, G.T, Thanassi, D.G, Li, H. | | Deposit date: | 2018-07-09 | | Release date: | 2018-10-17 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Handover mechanism of the growing pilus by the bacterial outer-membrane usher FimD.
Nature, 562, 2018
|
|
6E16
 
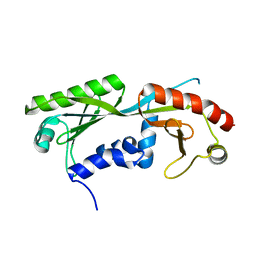 | | Ternary structure of c-Myc-TBP-TAF1 | | Descriptor: | Transcription initiation factor TFIID subunit 1,Myc proto-oncogene protein,TATA-box-binding protein | | Authors: | Wei, Y, Dong, A, Sunnerhagen, M, Penn, L, Tong, Y, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-07-09 | | Release date: | 2019-10-02 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Multiple direct interactions of TBP with the MYC oncoprotein.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6E17
 
 | |
6E18
 
 | | Crystal structure of Chlamydomonas reinhardtii HAP2 ectodomain provides structural insights of functional loops in green algae. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Baquero, E, Legrand, P, Rey, F.A. | | Deposit date: | 2018-07-09 | | Release date: | 2018-11-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Species-Specific Functional Regions of the Green Alga Gamete Fusion Protein HAP2 Revealed by Structural Studies.
Structure, 27, 2019
|
|
6E1A
 
 | | Menin bound to M-89 | | Descriptor: | (1S,2R)-2-[(4S)-2-methyl-4-{1-[(1-{4-[(pyridin-4-yl)sulfonyl]phenyl}azetidin-3-yl)methyl]piperidin-4-yl}-1,2,3,4-tetrahydroisoquinolin-4-yl]cyclopentyl methylcarbamate, Menin, praseodymium triacetate | | Authors: | Stuckey, J.A. | | Deposit date: | 2018-07-09 | | Release date: | 2019-07-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-Based Discovery of M-89 as a Highly Potent Inhibitor of the Menin-Mixed Lineage Leukemia (Menin-MLL) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6E1C
 
 | |
6E1F
 
 | |
