4IKW
 
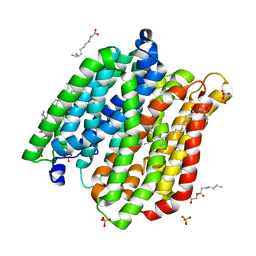 | | Crystal structure of peptide transporter POT in complex with sulfate | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Di-tripeptide ABC transporter (Permease), OLEIC ACID, ... | | Authors: | Doki, S, Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2012-12-28 | | Release date: | 2013-07-10 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.004 Å) | | Cite: | Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IKV
 
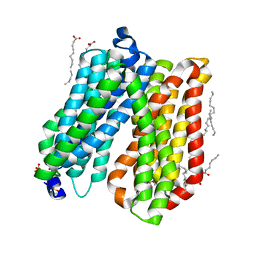 | | Crystal structure of peptide transporter POT | | Descriptor: | (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Di-tripeptide ABC transporter (Permease), OLEIC ACID, ... | | Authors: | Doki, S, Kato, H.E, Ishitani, R, Nureki, O. | | Deposit date: | 2012-12-28 | | Release date: | 2013-07-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for dynamic mechanism of proton-coupled symport by the peptide transporter POT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4IKE
 
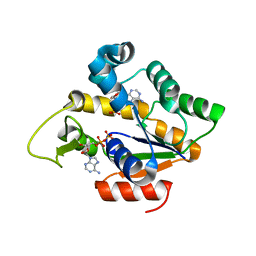 | |
4IK9
 
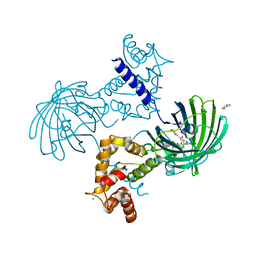 | | High resolution structure of GCaMP3 dimer form 2 at pH 7.5 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, RCaMP, ... | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK8
 
 | | High resolution structure of GCaMP3 dimer form 1 at pH 7.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK5
 
 | | High resolution structure of Delta-REST-GCaMP3 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK4
 
 | | High resolution structure of GCaMP3 at pH 5.0 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK3
 
 | | High resolution structure of GCaMP3 at pH 8.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.007 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4IK1
 
 | | High resolution structure of GCaMPJ at pH 8.5 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-02-05 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
4HVF
 
 | |
4H48
 
 | | 1.45 angstrom CyPet Structure at pH7.0 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Green fluorescent protein | | Authors: | Hu, X.-J, Liu, R. | | Deposit date: | 2012-09-17 | | Release date: | 2013-09-18 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure insight of the fluorescent state of CyPet
To be Published
|
|
4H47
 
 | | 1.9 angstrom CyPet structure at pH5.2 | | Descriptor: | ACETATE ION, Green fluorescent protein, SULFATE ION | | Authors: | Hu, X.-J, Liu, R. | | Deposit date: | 2012-09-17 | | Release date: | 2013-09-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights of the fluorescent states of CyPet
To be Published
|
|
4GPB
 
 | |
4GOB
 
 | | Low pH Crystal Structure of a reconstructed Kaede-type Red Fluorescent Protein, Least Evolved Ancestor (LEA) | | Descriptor: | Kaede-type Fluorescent Protein | | Authors: | Kim, H, Grunkemeyer, T.J, Chen, L, Fromme, R, Wachter, R.M. | | Deposit date: | 2012-08-19 | | Release date: | 2013-07-31 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Acid-base catalysis and crystal structures of a least evolved ancestral GFP-like protein undergoing green-to-red photoconversion.
Biochemistry, 52, 2013
|
|
4GFP
 
 | | 2.7 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in a second conformational state | | Descriptor: | 3-phosphoshikimate 1-carboxyvinyltransferase, BETA-MERCAPTOETHANOL | | Authors: | Light, S.H, Minasov, G, Krishna, S.N, Shuvalova, L, Papazisi, L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 2.7 Angstrom resolution structure of 3-phosphoshikimate 1-carboxyvinyltransferase (AroA) from Coxiella burnetii in second conformational state
TO BE PUBLISHED
|
|
4GF6
 
 | | crystal structure of GFP with cuprum bound at the Incorporated metal Chelating Amino Acid PYZ151 | | Descriptor: | CALCIUM ION, COPPER (II) ION, green fluorescent protein | | Authors: | Dong, J, Liu, X, Li, J, Wang, J, Gong, W. | | Deposit date: | 2012-08-03 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Genetic incorporation of a metal-chelating amino Acid as a probe for protein electron transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4GES
 
 | | crystal structure of GFP-TYR151PYZ with an unnatural amino acid incorporation | | Descriptor: | Green fluorescent protein | | Authors: | Dong, J, Liu, X, Li, J, Wang, J, Gong, W. | | Deposit date: | 2012-08-02 | | Release date: | 2012-08-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Genetic incorporation of a metal-chelating amino Acid as a probe for protein electron transfer.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
4FL6
 
 | | Crystal structure of the complex of the 3-MBT repeat domain of L3MBTL3 and UNC1215 | | Descriptor: | Lethal(3)malignant brain tumor-like protein 3, UNKNOWN ATOM OR ION, [2-(phenylamino)benzene-1,4-diyl]bis{[4-(pyrrolidin-1-yl)piperidin-1-yl]methanone} | | Authors: | Zhong, N, Tempel, W, Ravichandran, M, Dong, A, Ingerman, L.A, Graslund, S, Frye, S.V, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-14 | | Release date: | 2012-06-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Discovery of a chemical probe for the L3MBTL3 methyllysine reader domain.
Nat. Chem. Biol., 9, 2013
|
|
4EUL
 
 | | Crystal structure of enhanced Green Fluorescent Protein to 1.35A resolution reveals alternative conformations for Glu222 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, Green fluorescent protein, ... | | Authors: | Jones, D.D, Arpino, J.A.J, Rizkallah, P.J. | | Deposit date: | 2012-04-25 | | Release date: | 2012-10-03 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of enhanced green fluorescent protein to 1.35 a resolution reveals alternative conformations for glu222.
Plos One, 7, 2012
|
|
4EN1
 
 | |
4EMQ
 
 | | Crystal structure of a single mutant of Dronpa, the green-on-state PDM1-4 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, Fluorescent protein Dronpa, ... | | Authors: | Ngan, N.B, Van Hecke, K, Van Meervelt, L. | | Deposit date: | 2012-04-12 | | Release date: | 2012-11-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for the influence of a single mutation K145N on the oligomerization and photoswitching rate of Dronpa.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4EEU
 
 | | Crystal structure of phiLOV2.1 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.4068 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EET
 
 | | Crystal structure of iLOV | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EES
 
 | | Crystal structure of iLOV | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
4EER
 
 | | Crystal structure of LOV2 domain of Arabidopsis thaliana phototropin 2 C426A mutant | | Descriptor: | FLAVIN MONONUCLEOTIDE, Phototropin-2 | | Authors: | Hitomi, K, Christie, J.M, Arvai, A.S, Hartfield, K.A, Pratt, A.J, Tainer, J.A, Getzoff, E.D. | | Deposit date: | 2012-03-28 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.753 Å) | | Cite: | Structural Tuning of the Fluorescent Protein iLOV for Improved Photostability.
J.Biol.Chem., 287, 2012
|
|
