1S48
 
 | | Crystal structure of RNA-dependent RNA polymerase construct 1 (residues 71-679) from BVDV | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Choi, K.H, Groarke, J.M, Young, D.C, Kuhn, R.J, Smith, J.L, Pevear, D.C, Rossmann, M.G. | | Deposit date: | 2004-01-15 | | Release date: | 2004-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1K96
 
 | | CRYSTAL STRUCTURE OF CALCIUM BOUND HUMAN S100A6 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, S100A6 | | Authors: | Otterbein, L.R, Dominguez, R. | | Deposit date: | 2001-10-26 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Crystal structures of S100A6 in the Ca(2+)-free and Ca(2+)-bound states: the calcium sensor mechanism of S100 proteins revealed at atomic resolution.
Structure, 10, 2002
|
|
1K9K
 
 | | CRYSTAL STRUCTURE OF CALCIUM BOUND HUMAN S100A6 | | Descriptor: | BETA-MERCAPTOETHANOL, CALCIUM ION, S100A6 | | Authors: | Otterbein, L.R, Dominguez, R. | | Deposit date: | 2001-10-29 | | Release date: | 2002-04-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structures of S100A6 in the Ca(2+)-free and Ca(2+)-bound states: the calcium sensor mechanism of S100 proteins revealed at atomic resolution.
Structure, 10, 2002
|
|
1S4F
 
 | | Crystal Structure of RNA-dependent RNA polymerase construct 2 from bovine viral diarrhea virus (BVDV) | | Descriptor: | RNA-dependent RNA polymerase | | Authors: | Choi, K.H, Groarke, J.M, Young, D.C, Kuhn, R.J, Smith, J.L, Pevear, D.C, Rossmann, M.G. | | Deposit date: | 2004-01-16 | | Release date: | 2004-04-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1S70
 
 | | Complex between protein ser/thr phosphatase-1 (delta) and the myosin phosphatase targeting subunit 1 (MYPT1) | | Descriptor: | 130 kDa myosin-binding subunit of smooth muscle myosin phophatase (M130), MANGANESE (II) ION, Serine/threonine protein phosphatase PP1-beta (or delta) catalytic subunit, ... | | Authors: | Kerff, F, Terrak, M, Dominguez, R. | | Deposit date: | 2004-01-28 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of protein phosphatase 1 regulation
Nature, 429, 2004
|
|
1K78
 
 | | Pax5(1-149)+Ets-1(331-440)+DNA | | Descriptor: | C-ets-1 Protein, Paired Box Protein Pax5, Pax5/Ets Binding Site on the mb-1 promoter | | Authors: | Garvie, C.W, Hagman, J, Wolberger, C. | | Deposit date: | 2001-10-18 | | Release date: | 2002-01-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural studies of Ets-1/Pax5 complex formation on DNA.
Mol.Cell, 8, 2001
|
|
1S08
 
 | | Crystal Structure of the D147N Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1JB7
 
 | |
1S3K
 
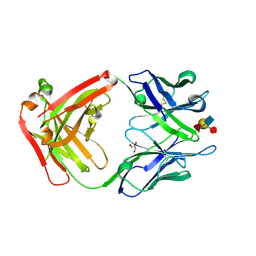 | | Crystal Structure of a Humanized Fab (hu3S193) in Complex with the Lewis Y Tetrasaccharide | | Descriptor: | GLYCEROL, HU3S193 Fab fragment, heavy chain, ... | | Authors: | Ramsland, P.A, Farrugia, W, Bradford, T.M, Hogarth, P.M, Scott, A.M. | | Deposit date: | 2004-01-13 | | Release date: | 2004-07-13 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural convergence of antibody binding of carbohydrate determinants in lewis y tumor antigens
J.Mol.Biol., 340, 2004
|
|
1JGQ
 
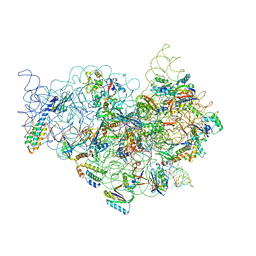 | | The Path of Messenger RNA Through the Ribosome. THIS FILE, 1JGQ, CONTAINS THE 30S RIBOSOME SUBUNIT, THREE TRNA, AND MRNA MOLECULES. 50S RIBOSOME SUBUNIT IS IN THE FILE 1GIY | | Descriptor: | 30S 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Yusupova, G.Z, Yusupov, M.M, Cate, J.H.D, Noller, H.F. | | Deposit date: | 2001-06-26 | | Release date: | 2001-07-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | The path of messenger RNA through the ribosome.
Cell(Cambridge,Mass.), 106, 2001
|
|
1S58
 
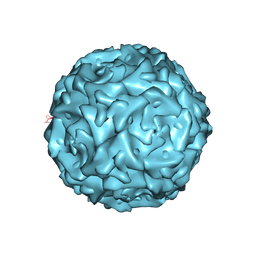 | |
1JA1
 
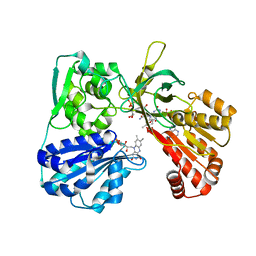 | | CYPOR-Triple Mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FLAVIN MONONUCLEOTIDE, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Hubbard, P.A, Shen, A.L, Paschke, R, Kasper, C.B, Kim, J.J. | | Deposit date: | 2001-05-29 | | Release date: | 2001-08-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | NADPH-cytochrome P450 oxidoreductase. Structural basis for hydride and electron transfer.
J.Biol.Chem., 276, 2001
|
|
1JBE
 
 | |
1S9X
 
 | | Crystal Structure Analysis of NY-ESO-1 epitope analogue, SLLMWITQA, in complex with HLA-A2 | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A-2 alpha chain, ... | | Authors: | Webb, A.I, Dunstone, M.A, Chen, W, Aguilar, M.I, Chen, Q, Chang, L, Kjer-Nielsen, L, Beddoe, T, McCluskey, J, Rossjohn, J, Purcell, A.W. | | Deposit date: | 2004-02-05 | | Release date: | 2004-09-28 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Functional and structural characteristics of NY-ESO-1-related HLA A2-restricted epitopes and the design of a novel immunogenic analogue
J.Biol.Chem., 279, 2004
|
|
1JCH
 
 | | Crystal Structure of Colicin E3 in Complex with its Immunity Protein | | Descriptor: | CITRIC ACID, COLICIN E3, COLICIN E3 IMMUNITY PROTEIN, ... | | Authors: | Soelaiman, S, Jakes, K, Wu, N, Li, C, Shoham, M. | | Deposit date: | 2001-06-09 | | Release date: | 2001-11-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.02 Å) | | Cite: | Crystal structure of colicin E3: implications for cell entry and ribosome inactivation.
Mol.Cell, 8, 2001
|
|
1S0A
 
 | | Crystal Structure of the Y17F Mutant of 7,8-Diaminopelargonic Acid Synthase | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, SODIUM ION | | Authors: | Sandmark, J, Eliot, A.C, Famm, K, Schneider, G, Kirsch, J.F. | | Deposit date: | 2003-12-30 | | Release date: | 2004-03-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Conserved and nonconserved residues in the substrate binding site of 7,8-diaminopelargonic acid synthase from Escherichia coli are essential for catalysis.
Biochemistry, 43, 2004
|
|
1JGP
 
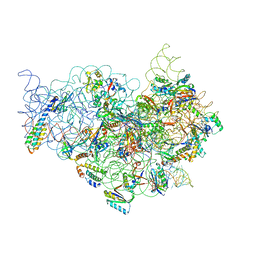 | | The Path of Messenger RNA Through the Ribosome. THIS FILE, 1JGP, CONTAINS THE 30S RIBOSOME SUBUNIT, THREE TRNA, AND MRNA MOLECULES. 50S RIBOSOME SUBUNIT IS IN THE FILE 1GIY | | Descriptor: | 30S 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Yusupova, G.Z, Yusupov, M.M, Cate, J.H.D, Noller, H.F. | | Deposit date: | 2001-06-26 | | Release date: | 2001-07-20 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (7 Å) | | Cite: | The path of messenger RNA through the ribosome.
Cell(Cambridge,Mass.), 106, 2001
|
|
5IAW
 
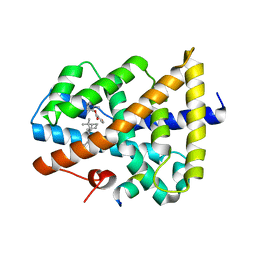 | | Novel natural FXR modulator with a unique binding mode | | Descriptor: | (1S,2R,4S)-1,7,7-trimethylbicyclo[2.2.1]heptan-2-yl 4-hydroxybenzoate, Bile acid receptor, Peptide from Nuclear receptor coactivator 2 | | Authors: | Lu, Y, Li, Y. | | Deposit date: | 2016-02-22 | | Release date: | 2017-03-08 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | A Novel Class of Natural FXR Modulators with a Unique Mode of Selective Co-regulator Assembly
Chembiochem, 18, 2017
|
|
1SFI
 
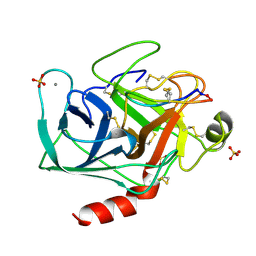 | | High resolution structure of a potent, cyclic protease inhibitor from sunflower seeds | | Descriptor: | CALCIUM ION, SULFATE ION, TRYPSIN, ... | | Authors: | Luckett, S, Garcia, R.S, Barker, J.J, Konarev, A.V, Shewry, P, Clarke, A.R, Brady, R.L. | | Deposit date: | 1998-12-16 | | Release date: | 1999-07-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | High-resolution structure of a potent, cyclic proteinase inhibitor from sunflower seeds.
J.Mol.Biol., 290, 1999
|
|
1RXO
 
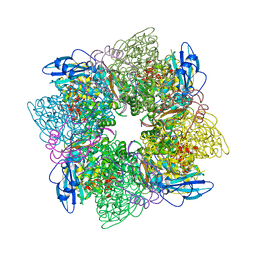 | | ACTIVATED SPINACH RUBISCO IN COMPLEX WITH ITS SUBSTRATE RIBULOSE-1,5-BISPHOSPHATE AND CALCIUM | | Descriptor: | CALCIUM ION, RIBULOSE BISPHOSPHATE CARBOXYLASE/OXYGENASE, RIBULOSE-1,5-DIPHOSPHATE | | Authors: | Taylor, T.C, Andersson, I. | | Deposit date: | 1996-12-06 | | Release date: | 1997-06-16 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The structure of the complex between rubisco and its natural substrate ribulose 1,5-bisphosphate.
J.Mol.Biol., 265, 1997
|
|
1KPI
 
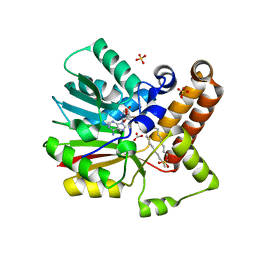 | | Crystal Structure of mycolic acid cyclopropane synthase CmaA2 complexed with SAH and DDDMAB | | Descriptor: | CARBONATE ION, CYCLOPROPANE-FATTY-ACYL-PHOSPHOLIPID SYNTHASE 2, DIDECYL-DIMETHYL-AMMONIUM, ... | | Authors: | Huang, C.-C, Smith, C.V, Jacobs Jr, W.R, Glickman, M.S, Sacchettini, J.C, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2001-12-30 | | Release date: | 2002-01-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structures of mycolic acid cyclopropane synthases from Mycobacterium tuberculosis
J.Biol.Chem., 277, 2002
|
|
1KHY
 
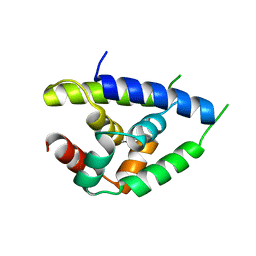 | |
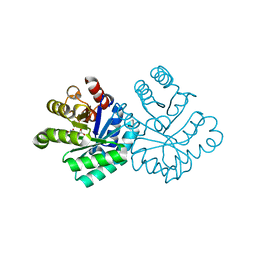
1KM5
 
 | |
1KNL
 
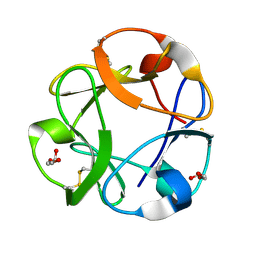 | | Streptomyces lividans Xylan Binding Domain cbm13 | | Descriptor: | ENDO-1,4-BETA-XYLANASE A, GLYCEROL | | Authors: | Notenboom, V, Boraston, A.B, Williams, S.J, Kilburn, D.G, Rose, D.R. | | Deposit date: | 2001-12-19 | | Release date: | 2002-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | High-resolution crystal structures of the lectin-like xylan binding domain from Streptomyces lividans xylanase 10A with bound substrates reveal a novel mode of xylan binding.
Biochemistry, 41, 2002
|
|
1K9M
 
 | | Co-crystal structure of tylosin bound to the 50S ribosomal subunit of Haloarcula marismortui | | Descriptor: | 23S RRNA, 5S RRNA, CADMIUM ION, ... | | Authors: | Hansen, J.L, Ippolito, J.A, Ban, N, Nissen, P, Moore, P.B, Steitz, T.A. | | Deposit date: | 2001-10-29 | | Release date: | 2002-07-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The structures of four macrolide antibiotics bound to the large ribosomal subunit.
Mol.Cell, 10, 2002
|
|
