1UX2
 
 | | X-ray structure of acetylcholine binding protein (AChBP) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETYLCHOLINE BINDING PROTEIN, ... | | Authors: | Celie, P.H.N, Van Rossum-fikkert, S.E, Van Dijk, W.J, Brejc, K, Smit, A.B, Sixma, T.K. | | Deposit date: | 2004-02-18 | | Release date: | 2004-03-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Nicotine and Carbamylcholine Binding to Nicotinic Acetylcholine Receptors as Studied in Achbp Crystal Structures
Neuron, 41, 2004
|
|
8BQF
 
 | | Adenylate Kinase L107I MUTANT | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE | | Authors: | Scheerer, D, Adkar, B.V, Bhattacharyya, S, Levy, D, Iljina, M, Iljina, I, Dym, O, Haran, G, Shakhnovich, E.I. | | Deposit date: | 2022-11-21 | | Release date: | 2023-05-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Allosteric communication between ligand binding domains modulates substrate inhibition in adenylate kinase.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3TQT
 
 | | Structure of the D-alanine-D-alanine ligase from Coxiella burnetii | | Descriptor: | D-alanine--D-alanine ligase | | Authors: | Rudolph, M, Cheung, J, Franklin, M.C, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
5AJT
 
 | | Crystal structure of ligand-free phosphoribohydrolase lonely guy from Claviceps purpurea | | Descriptor: | 1,2-ETHANEDIOL, D(-)-TARTARIC ACID, PHOSPHORIBOHYDROLASE LONELY GUY | | Authors: | Dzurova, L, Savino, S, Forneris, F. | | Deposit date: | 2015-02-27 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The three-dimensional structure of "Lonely Guy" from Claviceps purpurea provides insights into the phosphoribohydrolase function of Rossmann fold-containing lysine decarboxylase-like proteins.
Proteins, 83, 2015
|
|
7M9O
 
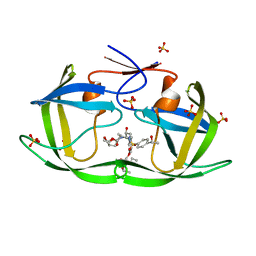 | | HIV-1 Protease (I84V) in Complex with LR3-48 | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-[{4-[(1R)-1-hydroxyethyl]benzene-1-sulfonyl}(2-methylpropyl)amino]butyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.895 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
7M9K
 
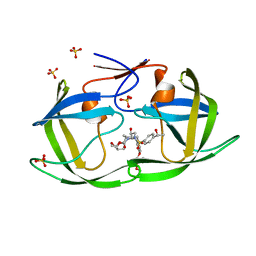 | | HIV-1 Protease WT (NL4-3) in Complex with LR3-48 | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-[{4-[(1R)-1-hydroxyethyl]benzene-1-sulfonyl}(2-methylpropyl)amino]butyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.838 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
7M9M
 
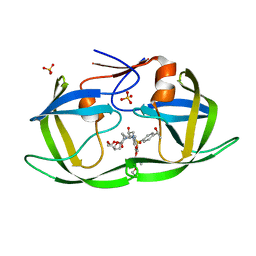 | | HIV-1 Protease WT (NL4-3) in Complex with LR3-55 | | Descriptor: | Protease, SULFATE ION, diethyl ({4-[(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-({4-[(1S)-1-hydroxyethyl]benzene-1-sulfonyl}[(2S)-2-methylbutyl]amino)butyl]phenoxy}methyl)phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.881 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
7MAJ
 
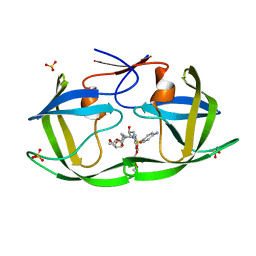 | | HIV-1 Protease (I84V) in Complex with PU6 (LR3-66) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-{[(4-aminophenyl)sulfonyl](2-ethylbutyl)amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.871 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
7MAK
 
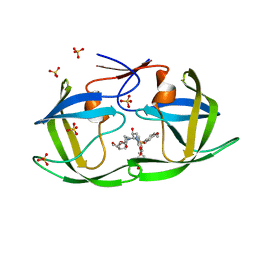 | | HIV-1 Protease (I84V) in Complex with PU7 (LR3-67) | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-4-{(2-ethylbutyl)[(4-methoxyphenyl)sulfonyl]amino}-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxybutyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.972 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
7MAG
 
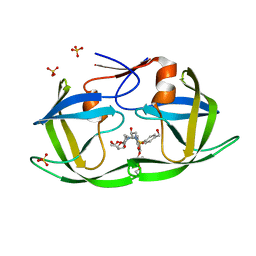 | | HIV-1 Protease (I84V) in Complex with PU3 (LR3-69) | | Descriptor: | Protease, SULFATE ION, diethyl ({4-[(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-({[4-(hydroxymethyl)phenyl]sulfonyl}[(2S)-2-methylbutyl]amino)butyl]phenoxy}methyl)phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
7M9P
 
 | | HIV-1 Protease (I84V) in Complex with LR3-55 | | Descriptor: | Protease, SULFATE ION, diethyl ({4-[(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-({4-[(1S)-1-hydroxyethyl]benzene-1-sulfonyl}[(2S)-2-methylbutyl]amino)butyl]phenoxy}methyl)phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.819 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
7M9N
 
 | | HIV-1 Protease (I84V) in Complex with LR3-68 | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-[{4-[(1S)-1-hydroxyethyl]benzene-1-sulfonyl}(2-methylpropyl)amino]butyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-20 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
7M9J
 
 | | HIV-1 Protease WT (NL4-3) in Complex with LR3-68 | | Descriptor: | Protease, SULFATE ION, diethyl [(4-{(2S,3R)-2-[({[(3R,3aS,6aR)-hexahydrofuro[2,3-b]furan-3-yl]oxy}carbonyl)amino]-3-hydroxy-4-[{4-[(1S)-1-hydroxyethyl]benzene-1-sulfonyl}(2-methylpropyl)amino]butyl}phenoxy)methyl]phosphonate | | Authors: | Lockbaum, G.J, Schiffer, C.A. | | Deposit date: | 2021-03-31 | | Release date: | 2022-04-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | HIV-1 Protease in Complex with ligands
To Be Published
|
|
7RGW
 
 | |
1QUI
 
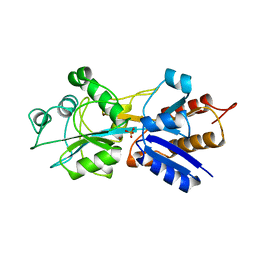 | | PHOSPHATE-BINDING PROTEIN MUTANT WITH ASP 137 REPLACED BY GLY COMPLEX WITH BROMINE AND PHOSPHATE | | Descriptor: | BROMIDE ION, PHOSPHATE ION, PHOSPHATE-BINDING PROTEIN | | Authors: | Yao, N, Choudhary, A, Ledvina, P.S, Quiocho, F.A. | | Deposit date: | 1995-11-11 | | Release date: | 1996-07-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Modulation of a salt link does not affect binding of phosphate to its specific active transport receptor.
Biochemistry, 35, 1996
|
|
2Q3L
 
 | |
4XB9
 
 | |
5EKB
 
 | |
6LR2
 
 | |
3TGU
 
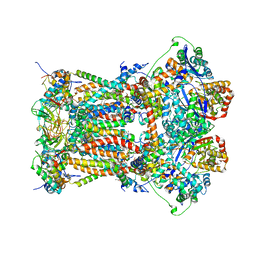 | | Cytochrome bc1 complex from chicken with pfvs-designed moa inhibitor bound | | Descriptor: | 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine, CARDIOLIPIN, Coenzyme Q10, ... | | Authors: | Huang, L.-S, Yang, G.-F, Berry, E.A. | | Deposit date: | 2011-08-17 | | Release date: | 2012-07-04 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Computational discovery of picomolar Q(o) site inhibitors of cytochrome bc1 complex.
J.Am.Chem.Soc., 134, 2012
|
|
4ZYC
 
 | |
2FZP
 
 | | Crystal structure of the USP8 interaction domain of human NRDP1 | | Descriptor: | ring finger protein 41 isoform 1 | | Authors: | Walker, J.R, Avvakumov, G.V, Xue, S, Newman, E.M, Butler-Cole, C, Finerty Jr, P.J, Weigelt, J, Sundstrom, M, Arrowsmith, C, Edwards, A, Bochkarev, A, Dhe-Paganon, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-02-10 | | Release date: | 2006-03-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Amino-terminal Dimerization, NRDP1-Rhodanese Interaction, and Inhibited Catalytic Domain Conformation of the Ubiquitin-specific Protease 8 (USP8).
J.Biol.Chem., 281, 2006
|
|
6USV
 
 | | Crystal structure of GluN1/GluN2A ligand-binding domain in complex with glycine and SDZ 220-040 | | Descriptor: | (2S)-2-amino-3-[2',4'-dichloro-4-hydroxy-5-(phosphonomethyl)biphenyl-3-yl]propanoic acid, GLYCEROL, GLYCINE, ... | | Authors: | Romero-Hernandez, A, Tajima, N, Chou, T, Furukawa, h. | | Deposit date: | 2019-10-28 | | Release date: | 2020-07-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Structural Basis of Functional Transitions in Mammalian NMDA Receptors.
Cell, 182, 2020
|
|
6XKD
 
 | | Structure of ligand-bound mouse cGAMP hydrolase ENPP1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Fernandez, D, Li, L. | | Deposit date: | 2020-06-26 | | Release date: | 2021-06-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structure-Aided Development of Small-Molecule Inhibitors of ENPP1, the Extracellular Phosphodiesterase of the Immunotransmitter cGAMP.
Cell Chem Biol, 27, 2020
|
|
4XBW
 
 | |
