3D90
 
 | | Crystal structure of the human progesterone receptor ligand-binding domain bound to levonorgestrel | | Descriptor: | 13-BETA-ETHYL-17-ALPHA-ETHYNYL-17-BETA-HYDROXYGON-4-EN-3-ONE, Progesterone receptor | | Authors: | Petit-Topin, I, Turque, N, Ulman, A, Gainer, E, Rafestin-Oblin, M.E, Fagart, J. | | Deposit date: | 2008-05-26 | | Release date: | 2009-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Met909 plays a key role in the activation of the progesterone receptor and also in the high potency of 13-ethyl progestins
Mol.Pharmacol., 75, 2009
|
|
3D9U
 
 | |
4KCA
 
 | | Crystal Structure of Endo-1,5-alpha-L-arabinanase from a Bovine Ruminal Metagenomic Library | | Descriptor: | Endo-1,5-alpha-L-arabinanase, GLYCEROL, IODIDE ION, ... | | Authors: | Santos, C.R, Polo, C.C, Costa, M.C.M.F, Nascimento, A.F.Z, Wong, D.W.S, Murakami, M.T. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanistic strategies for catalysis adopted by evolutionary distinct family 43 arabinanases.
J.Biol.Chem., 289, 2014
|
|
4UBW
 
 | |
3DA0
 
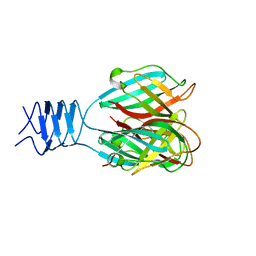 | | Crystal structure of a cleaved form of a chimeric receptor binding protein from Lactococcal phages subspecies TP901-1 and p2 | | Descriptor: | Cleaved chimeric receptor binding protein from bacteriophages TP901-1 and p2 | | Authors: | Siponen, M.I, Blangy, S, Spinelli, S, Vera, L, Cambillau, C, Campanacci, V. | | Deposit date: | 2008-05-28 | | Release date: | 2009-06-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a chimeric receptor binding protein constructed from two lactococcal phages.
J.Bacteriol., 191, 2009
|
|
3M1W
 
 | |
4KCR
 
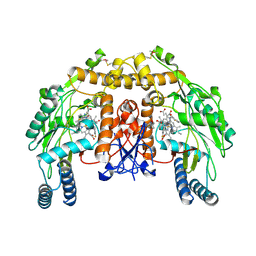 | | Structure of bovine endothelial nitric oxide synthase heme domain in complex with N-(3-(((3-fluorophenethyl)amino)methyl)phenyl)thiophene-2-carboximidamide | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2013-04-24 | | Release date: | 2014-02-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Potent and Selective Double-Headed Thiophene-2-carboximidamide Inhibitors of Neuronal Nitric Oxide Synthase for the Treatment of Melanoma.
J.Med.Chem., 57, 2014
|
|
3M29
 
 | |
4KDI
 
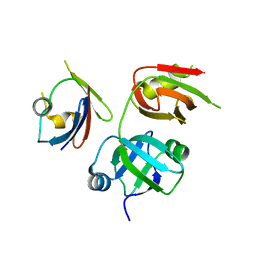 | | Crystal structure of p97/VCP N in complex with OTU1 UBXL | | Descriptor: | Transitional endoplasmic reticulum ATPase, Ubiquitin thioesterase OTU1 | | Authors: | Kim, S.J, Kim, E.E. | | Deposit date: | 2013-04-25 | | Release date: | 2014-03-19 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural Basis for Ovarian Tumor Domain-containing Protein 1 (OTU1) Binding to p97/Valosin-containing Protein (VCP).
J.Biol.Chem., 289, 2014
|
|
3DA8
 
 | | Crystal structure of PurN from Mycobacterium tuberculosis | | Descriptor: | BETA-MERCAPTOETHANOL, IODIDE ION, MAGNESIUM ION, ... | | Authors: | Zhang, Z, Squire, C.J, Baker, E.N. | | Deposit date: | 2008-05-28 | | Release date: | 2009-05-12 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structures of glycinamide ribonucleotide transformylase (PurN) from Mycobacterium tuberculosis reveal a novel dimer with relevance to drug discovery.
J.Mol.Biol., 389, 2009
|
|
3MIB
 
 | | Oxidized (Cu2+) peptidylglycine alpha-hydroxylating monooxygenase (PHM) with bound nitrite | | Descriptor: | COPPER (II) ION, GLYCEROL, NICKEL (II) ION, ... | | Authors: | Chufan, E.E, Eipper, B.A, Mains, R.E, Amzel, L.M. | | Deposit date: | 2010-04-10 | | Release date: | 2010-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Differential Reactivity Between the Two Copper Sites of Peptidylglycine alpha-Hydroxylating Monooxygenase (PHM)
J.Am.Chem.Soc., 132, 2010
|
|
4KPF
 
 | | Novel fluoroquinolones in complex with topoisomerase IV from S. pneumoniae and E-site G-gate | | Descriptor: | (3aS,4R)-4-amino-13-cyclopropyl-8-fluoro-10-oxo-3a,4,5,6,10,13-hexahydro-1H,3H-pyrrolo[2',1':3,4][1,4]oxazepino[5,6-h]quinoline-11-carboxylic acid, E-site1, E-site2, ... | | Authors: | Laponogov, I, Pan, X.-S, Vesekov, D.A, Cirz, R.T, Wagman, A.S, Moser, H.E, Fisher, L.M, Sanderson, M.R. | | Deposit date: | 2013-05-13 | | Release date: | 2014-11-26 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Exploring the active site of the Streptococcus pneumoniae topoisomerase IV-DNA cleavage complex with novel 7,8-bridged fluoroquinolones.
Open Biol, 6, 2016
|
|
3MJT
 
 | | Structure of A-type Ketoreductases from Modular Polyketide Synthase | | Descriptor: | AmphB, GLYCEROL, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zheng, J, Taylor, C.A, Piasecki, S.K, Keatinge-Clay, A.T. | | Deposit date: | 2010-04-13 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and Functional Analysis of A-Type Ketoreductases from the Amphotericin Modular Polyketide Synthase.
Structure, 18, 2010
|
|
4KPO
 
 | | Plant nucleoside hydrolase - ZmNRh3 enzyme | | Descriptor: | CALCIUM ION, Nucleoside N-ribohydrolase 3 | | Authors: | Morera, S, Vigouroux, A, Kopecny, D. | | Deposit date: | 2013-05-14 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structure and Function of Nucleoside Hydrolases from Physcomitrella patens and Maize Catalyzing the Hydrolysis of Purine, Pyrimidine, and Cytokinin Ribosides.
Plant Physiol., 163, 2013
|
|
4KPW
 
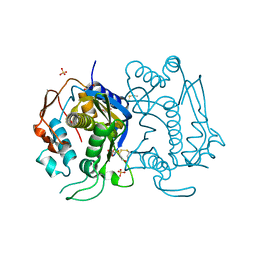 | |
3DAZ
 
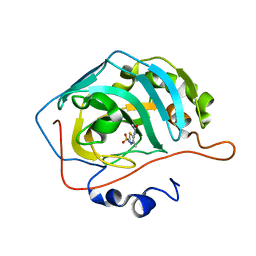 | | Use of Carbonic Anhydrase II, IX Active-Site Mimic, for the Purpose of Screening Inhibitors for Possible Anti-Cancer Properties | | Descriptor: | Carbonic anhydrase 2, N-(3-methyl-5-sulfamoyl-1,3,4-thiadiazol-2(3H)-ylidene)acetamide, ZINC ION | | Authors: | Genis, C, Sippel, K.H, Case, N, Govindasamy, L, Agbandje-Mckenna, M, Mckenna, R. | | Deposit date: | 2008-05-30 | | Release date: | 2009-03-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Design of a carbonic anhydrase IX active-site mimic to screen inhibitors for possible anticancer properties
Biochemistry, 48, 2009
|
|
3DBC
 
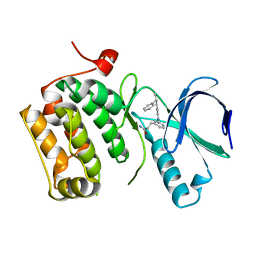 | | Crystal structure of an activated (Thr->Asp) Polo-like kinase 1 (Plk1) catalytic domain in complex with Compound 257 | | Descriptor: | 3-[3-(3-methyl-6-{[(1S)-1-phenylethyl]amino}-1H-pyrazolo[4,3-c]pyridin-1-yl)phenyl]propanamide, Polo-like kinase 1 | | Authors: | Elling, R.A, Zhu, J, Barr, K.J, Romanowski, M.J. | | Deposit date: | 2008-05-31 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Design and synthesis of 2-amino-pyrazolopyridines as Polo-like kinase 1 inhibitors.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3MKR
 
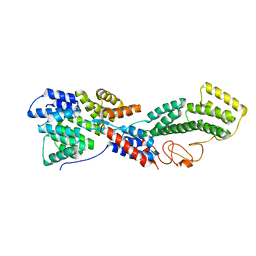 | |
4UH6
 
 | | Structure of human nNOS R354A G357D mutant heme domain in complex with 3-(2-(6-Amino-4-methylpyridin-2-yl)ethyl)-5-(methyl(2-(methylamino)ethyl)amino)benzonitrile | | Descriptor: | 3-(2-(6-AMINO-4-METHYLPYRIDIN-2-YL)ETHYL)-5-(METHYL(2-(METHYLAMINO)ETHYL)AMINO)BENZONITRILE, 5,6,7,8-TETRAHYDROBIOPTERIN, CHLORIDE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2015-03-23 | | Release date: | 2015-07-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | 2-Aminopyridines with a Truncated Side Chain to Improve Human Neuronal Nitric Oxide Synthase Inhibitory Potency and Selectivity.
J.Med.Chem., 58, 2015
|
|
3M2L
 
 | |
4KEG
 
 | | Crystal Structure of MBP Fused Human SPLUNC1 | | Descriptor: | MAGNESIUM ION, Maltose-binding periplasmic/Palate lung and nasal epithelium clone fusion protein, octyl beta-D-glucopyranoside | | Authors: | Ning, F, Wang, C, Niu, L, Chu, H.W, Zhang, G. | | Deposit date: | 2013-04-25 | | Release date: | 2014-04-30 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Lipid Ligands of the SPLUNC1 Protein
To be Published
|
|
4WE8
 
 | | The crystal structure of hemagglutinin of influenza virus A/Victoria/361/2011 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin | | Authors: | Yang, H, Carney, P.J, Chang, J.C, Guo, Z, Villanueva, J.M, Stevens, J. | | Deposit date: | 2014-09-09 | | Release date: | 2015-02-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and receptor binding preferences of recombinant human A(H3N2) virus hemagglutinins.
Virology, 477C, 2015
|
|
4KJK
 
 | | Room Temperature WT DHFR | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
3DBN
 
 | | Crystal structure of the Streptoccocus suis serotype2 D-mannonate dehydratase in complex with its substrate | | Descriptor: | D-MANNONIC ACID, MANGANESE (II) ION, Mannonate dehydratase | | Authors: | Peng, H, Zhang, Q, Gao, F, Gao, G.F. | | Deposit date: | 2008-06-02 | | Release date: | 2009-06-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of Streptococcus suis mannonate dehydratase (ManD) and its complex with substrate: genetic and biochemical evidence for a catalytic mechanism
J.Bacteriol., 191, 2009
|
|
3M3T
 
 | |
