4TW3
 
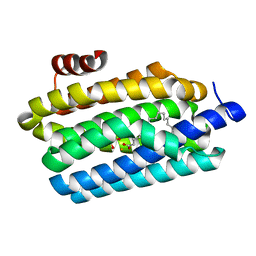 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde decarbonylase, FE (III) ION, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-06-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
4TZ8
 
 | | Structure of human ATAD2 bromodomain bound to fragment inhibitor | | Descriptor: | 2-amino-7,7-dimethyl-5,6,7,8-tetrahydro-4H-[1,3]thiazolo[5,4-c]azepin-4-one, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Harner, M.J, Chauder, B.A, Phan, J, Fesik, S.W. | | Deposit date: | 2014-07-09 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Screening of the Bromodomain of ATAD2.
J.Med.Chem., 57, 2014
|
|
4KQQ
 
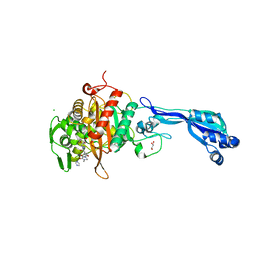 | | CRYSTAL STRUCTURE OF PENICILLIN-BINDING PROTEIN 3 FROM PSEUDOMONAS AERUGINOSA IN COMPLEX WITH (5S)-Penicilloic Acid | | Descriptor: | (2S,4S)-2-[(R)-carboxy{[(2R)-2-{[(4-ethyl-2,3-dioxopiperazin-1-yl)carbonyl]amino}-2-phenylacetyl]amino}methyl]-5,5-dimethyl-1,3-thiazolidine-4-carboxylic acid, CHLORIDE ION, GLYCEROL, ... | | Authors: | Nettleship, J.E, Stuart, D.I, Owens, R.J, Ren, J. | | Deposit date: | 2013-05-15 | | Release date: | 2013-11-06 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Binding of (5S)-Penicilloic Acid to Penicillin Binding Protein 3.
Acs Chem.Biol., 8, 2013
|
|
8CNN
 
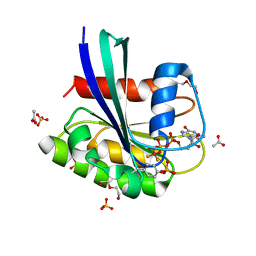 | | BeF3 Phospho-HRas GSA complex | | Descriptor: | ACETATE ION, BERYLLIUM TRIFLUORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Baumann, P, Jin, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Far-reaching effects of tyrosine64 phosphorylation on Ras revealed with BeF 3 - complexes.
Commun Chem, 7, 2024
|
|
8CNJ
 
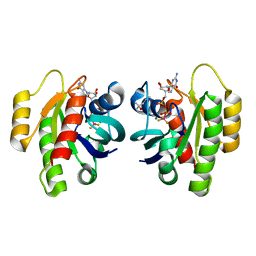 | | HRas(1-166) in complex with GDP and BeF3- | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BERYLLIUM TRIFLUORIDE ION, GTPase HRas, ... | | Authors: | Baumann, P, Jin, Y. | | Deposit date: | 2023-02-23 | | Release date: | 2023-09-27 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Far-reaching effects of tyrosine64 phosphorylation on Ras revealed with BeF 3 - complexes.
Commun Chem, 7, 2024
|
|
6ZVO
 
 | | Crystal structure of unliganded MLKL executioner domain | | Descriptor: | BROMIDE ION, CHLORIDE ION, Mixed lineage kinase domain-like protein | | Authors: | Fiegen, D, Bauer, M, Nar, H. | | Deposit date: | 2020-07-27 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.371 Å) | | Cite: | Locking mixed-lineage kinase domain-like protein in its auto-inhibited state prevents necroptosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6ZZ1
 
 | | Crystal structure of MLKL executioner domain in complex with a covalent inhibitor | | Descriptor: | 7-(2-methoxyethoxymethyl)-1,3-dimethyl-purine-2,6-dione, Mixed lineage kinase domain-like protein | | Authors: | Fiegen, D, Bauer, M, Nar, H. | | Deposit date: | 2020-08-03 | | Release date: | 2020-12-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Locking mixed-lineage kinase domain-like protein in its auto-inhibited state prevents necroptosis.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
5A7U
 
 | | Single-particle cryo-EM of co-translational folded adr1 domain inside the E. coli ribosome exit tunnel. | | Descriptor: | REGULATORY PROTEIN ADR1, ZINC ION | | Authors: | Nilsson, O.B, Hedman, R, Marino, J, Wickles, S, Bischoff, L, Johansson, M, Muller-Lucks, A, Trovato, F, Puglisi, J.D, O'Brien, E, Beckmann, R, von Heijne, G. | | Deposit date: | 2015-07-10 | | Release date: | 2015-09-16 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Cotranslational Protein Folding Inside the Ribosome Exit Tunnel.
Cell Rep., 12, 2015
|
|
3ZS2
 
 | | TyrB25,NMePheB26,LysB28,ProB29-insulin analogue crystal structure | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Antolikova, E, Zakova, L, Turkenburg, J.P, Watson, C.J, Hanclova, I, Sanda, M, Cooper, A, Kraus, T, Brzozowski, A.M, Jiracek, J.A. | | Deposit date: | 2011-06-21 | | Release date: | 2011-08-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Non-Equivalent Role of Inter- and Intramolecular Hydrogen Bonds in the Insulin Dimer Interface.
J.Biol.Chem., 286, 2011
|
|
3ZQR
 
 | | NMePheB25 insulin analogue crystal structure | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Antolikova, E, Zakova, L, Turkenburg, J.P, Watson, C.J, Hanclova, I, Sanda, M, Cooper, A, Kraus, T, Brzozowski, A.M, Jiracek, J.A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-08-24 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Non-Equivalent Role of Inter- and Intramolecular Hydrogen Bonds in the Insulin Dimer Interface.
J.Biol.Chem., 286, 2011
|
|
3ZU1
 
 | |
4AJX
 
 | | Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin | | Descriptor: | IMIDAZOLE, INSULIN, N-(16-Carboxyhexadecanoyl)-L-glutamic acid, ... | | Authors: | Steensgaard, D.B, Schluckebier, G, Strauss, H.M, Norrman, M, Thomsen, J.K, Friderichsen, A.V, Havelund, S, Jonassen, I. | | Deposit date: | 2012-02-20 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Ligand Controlled Assembly of Hexamers, Dihexamers, and Linear Multihexamer Structures by the Engineered Acylated Insulin Degludec.
Biochemistry, 52, 2013
|
|
4AKJ
 
 | | Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Steensgaard, D.B, Schluckebier, G, Strauss, H.M, Norrman, M, Thomsen, J.K, Friderichsen, A.V, Havelund, S, Jonassen, I. | | Deposit date: | 2012-02-23 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Ligand Controlled Assembly of Hexamers, Dihexamers, and Linear Multihexamer Structures by the Engineered Acylated Insulin Degludec.
Biochemistry, 52, 2013
|
|
4AJZ
 
 | | Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin | | Descriptor: | CHLORIDE ION, INSULIN A CHAIN, INSULIN B CHAIN, ... | | Authors: | Steensgaard, D.B, Schluckebier, G, Strauss, H.M, Norrman, M, Thomsen, J.K, Friderichsen, A.V, Havelund, S, Jonassen, I. | | Deposit date: | 2012-02-21 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand Controlled Assembly of Hexamers, Dihexamers, and Linear Multihexamer Structures by the Engineered Acylated Insulin Degludec.
Biochemistry, 52, 2013
|
|
4AK0
 
 | | Ligand controlled assembly of hexamers, dihexamers, and linear multihexamer structures by an engineered acylated insulin | | Descriptor: | INSULIN A CHAIN, INSULIN B CHAIN | | Authors: | Steensgaard, D.B, Schluckebier, G, Strauss, H.M, Norrman, M, Thomsen, J.K, Friderichsen, A.V, Havelund, S, Jonassen, I. | | Deposit date: | 2012-02-21 | | Release date: | 2013-01-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Ligand Controlled Assembly of Hexamers, Dihexamers, and Linear Multihexamer Structures by the Engineered Acylated Insulin Degludec.
Biochemistry, 52, 2013
|
|
8KG3
 
 | | Structure of THOUSAND-GRAIN WEIGHT 6 (TGW6) | | Descriptor: | Os06g0623700 protein | | Authors: | Akabane, T, Suzuki, N, Matsumura, H, Yoshizawa, T, Tsuchiya, W, Katoh, E, Hirotsu, N. | | Deposit date: | 2023-08-17 | | Release date: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | THOUSAND-GRAIN WEIGHT 6, which is an IAA-glucose hydrolase, preferentially recognizes the structure of the indole ring.
Sci Rep, 14, 2024
|
|
7TTV
 
 | |
8OPI
 
 | | VHH Z70 mutant 1 in interaction with PHF6 Tau peptide | | Descriptor: | PHF6 Tau peptide, VHH Z70 mutant 1 | | Authors: | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | Deposit date: | 2023-04-07 | | Release date: | 2023-05-31 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
8OP0
 
 | | VHH Z70 mutant 20 in interaction with PHF6 Tau peptide | | Descriptor: | Tau PHF6 peptide, VHH Z70 mutant 20 | | Authors: | Dupre, E, Mortelecque, J, NGuyen, M, Hanoulle, X, Landrieu, I. | | Deposit date: | 2023-04-06 | | Release date: | 2023-05-31 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | A selection and optimization strategy for single-domain antibodies targeting the PHF6 linear peptide within the tau intrinsically disordered protein.
J.Biol.Chem., 300, 2024
|
|
5AZQ
 
 | | Crystal structure of cyano-cobalt(III) tetradehydrocorrin in the heme pocket of horse heart myoglobin | | Descriptor: | (1R,19R) cobalt tetradehydrocorrin, (1S,19S) cobalt tetradehydrocorrin, CYANIDE ION, ... | | Authors: | Mizohata, E, Morita, Y, Oohora, K, Inoue, T, Hayashi, T. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal Structures and Coordination Behavior of Aqua- and Cyano-Co(III) Tetradehydrocorrins in the Heme Pocket of Myoglobin
Inorg.Chem., 55, 2016
|
|
5AZR
 
 | | Crystal structure of aqua-cobalt(III) tetradehydrocorrin in the heme pocket of horse heart myoglobin | | Descriptor: | (1R,19R) cobalt tetradehydrocorrin, (1S,19S) cobalt tetradehydrocorrin, GLYCEROL, ... | | Authors: | Mizohata, E, Morita, Y, Oohora, K, Inoue, T, Hayashi, T. | | Deposit date: | 2015-10-21 | | Release date: | 2016-01-20 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Crystal Structures and Coordination Behavior of Aqua- and Cyano-Co(III) Tetradehydrocorrins in the Heme Pocket of Myoglobin
Inorg.Chem., 55, 2016
|
|
5CTU
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with a fragment | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-(thiophen-2-yl)thieno[2,3-d]pyrimidin-4(1H)-one, CHLORIDE ION, ... | | Authors: | Andersen, O.A, Barker, J, Cheng, R.K, Kahmann, J, Felicetti, B, Wood, M, Scheich, C, Mesleh, M, Cross, J.B, Zhang, J, Yang, Q, Lippa, B, Ryan, M.D. | | Deposit date: | 2015-07-24 | | Release date: | 2016-02-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Fragment-based discovery of DNA gyrase inhibitors targeting the ATPase subunit of GyrB.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5DDO
 
 | |
5CPV
 
 | |
5DDP
 
 | | L-glutamine riboswitch bound with L-glutamine | | Descriptor: | GLUTAMINE, MAGNESIUM ION, RNA (61-MER), ... | | Authors: | Ren, A, Patel, D.J. | | Deposit date: | 2015-08-25 | | Release date: | 2015-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | Structural and Dynamic Basis for Low-Affinity, High-Selectivity Binding of L-Glutamine by the Glutamine Riboswitch.
Cell Rep, 13, 2015
|
|
