8PHL
 
 | | Human carbonic anhydrase II containing 4-fluorophenylalanine | | Descriptor: | Carbonic anhydrase 2, MERCURIBENZOIC ACID, ZINC ION | | Authors: | Pham, L.B.T, Costantino, A, Barbieri, L, Calderone, V, Luchinat, E, Banci, L. | | Deposit date: | 2023-06-20 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Controlling the incorporation of fluorinated amino acids in human cells and its structural impact.
Protein Sci., 33, 2024
|
|
7R0X
 
 | |
8UUF
 
 | | SARS-CoV-2 papain-like protease (PLpro) with inhibitor Jun11941 | | Descriptor: | ACETATE ION, CHLORIDE ION, N-{(1R)-1-[(3M,5P)-3,5-bis(1-methyl-1H-pyrazol-4-yl)phenyl]ethyl}-5-[2-(dimethylamino)ethoxy]-2-methylbenzamide, ... | | Authors: | Ansari, A, Tan, B, Riuz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-01 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
8UUY
 
 | | SARS-CoV-2 papain-like protease (PLpro) complex with inhibitor Jun12129 | | Descriptor: | ACETATE ION, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
6C9F
 
 | | AMP-activated protein kinase bound to pharmacological activator R734 | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1,5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Yan, Y, Zhou, X.E, Novick, S, Shaw, S.J, Li, Y, Hitoshi, Y, Brunzelle, J.S, Griffin, P.R, Xu, H.E, Melcher, K. | | Deposit date: | 2018-01-26 | | Release date: | 2018-11-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.924 Å) | | Cite: | Structures of AMP-activated protein kinase bound to novel pharmacological activators in phosphorylated, non-phosphorylated, and nucleotide-free states.
J. Biol. Chem., 294, 2019
|
|
5JAJ
 
 | |
7RNX
 
 | | CLC-ec1 at pH 7.5 100mM Cl Swap | | Descriptor: | H(+)/Cl(-) exchange transporter ClcA | | Authors: | Fortea, E, Boudker, O, Accardi, A. | | Deposit date: | 2021-07-30 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.42 Å) | | Cite: | Structural basis of common gate activation in CLC transporters
To Be Published
|
|
7RP6
 
 | |
8UQZ
 
 | | Round 18 Arylesterase Variant of Phosphotriesterase Bound to Gadolinium(III) Measured at 9.5 keV | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, GADOLINIUM ION, ... | | Authors: | Breeze, C.W, Frkic, R.L, Campbell, E.C, Jackson, C.J. | | Deposit date: | 2023-10-25 | | Release date: | 2024-04-03 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Mononuclear binding and catalytic activity of europium(III) and gadolinium(III) at the active site of the model metalloenzyme phosphotriesterase.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
6MLX
 
 | | Crystal structure of T. pallidum Leucine Rich Repeat protein (TpLRR) | | Descriptor: | Leucine-rich repeat protein TpLRR | | Authors: | Ramaswamy, R, Loveless, B.C, Houston, S, Cameron, C.E, Boulanger, M.J. | | Deposit date: | 2018-09-28 | | Release date: | 2019-07-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural characterization of Treponema pallidum Tp0225 reveals an unexpected leucine-rich repeat architecture.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
5JB2
 
 | |
8UUU
 
 | | SARS-Cov-2 papain-like protease (PLpro) with inhibitor Jun12162 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ansari, A, Tan, B, Ruiz, F.X, Arnold, E, Wang, J. | | Deposit date: | 2023-11-02 | | Release date: | 2024-04-03 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Design of a SARS-CoV-2 papain-like protease inhibitor with antiviral efficacy in a mouse model.
Science, 383, 2024
|
|
5MZJ
 
 | | Crystal structure of stabilized A2A adenosine receptor A2AR-StaR2-bRIL in complex with theophylline at 2.0A resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, Adenosine receptor A2a,Soluble cytochrome b562,Adenosine receptor A2a, CHOLESTEROL, ... | | Authors: | Cheng, R.K.Y, Segala, E, Robertson, N, Deflorian, F, Dore, A.S, Errey, J.C, Fiez-Vandal, C, Marshall, F.H, Cooke, R.M. | | Deposit date: | 2017-01-31 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of Human A1 and A2A Adenosine Receptors with Xanthines Reveal Determinants of Selectivity.
Structure, 25, 2017
|
|
7RO0
 
 | |
8BL3
 
 | |
6R8P
 
 | | Notum fragment 723 | | Descriptor: | 2-(2-methylphenoxy)-~{N}-pyridin-3-yl-ethanamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, DIMETHYL SULFOXIDE, ... | | Authors: | Zhao, Y, Jones, E.Y. | | Deposit date: | 2019-04-02 | | Release date: | 2019-05-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Discovery of 2-phenoxyacetamides as inhibitors of the Wnt-depalmitoleating enzyme NOTUM from an X-ray fragment screen.
Medchemcomm, 10, 2019
|
|
5N05
 
 | | X-ray crystal structure of an LPMO | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Auxiliary activity 9, CHLORIDE ION, ... | | Authors: | Frandsen, K.E.H, Poulsen, J.-C.N, Lo Leggio, L. | | Deposit date: | 2017-02-02 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Unliganded and substrate bound structures of the cellooligosaccharide active lytic polysaccharide monooxygenase LsAA9A at low pH.
Carbohydr. Res., 448, 2017
|
|
8V84
 
 | | 60S ribosome biogenesis intermediate (Dbp10 catalytic structure - Overall map) | | Descriptor: | 25S rRNA (cytosine(2870)-C(5))-methyltransferase, 60S ribosomal protein L13-A, 60S ribosomal protein L14-A, ... | | Authors: | Cruz, V.E, Weirich, C.S, Peddada, N, Erzberger, J.P. | | Deposit date: | 2023-12-04 | | Release date: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | The DEAD-box ATPase Dbp10/DDX54 initiates peptidyl transferase center formation during 60S ribosome biogenesis.
Nat Commun, 15, 2024
|
|
6RCP
 
 | |
6MH6
 
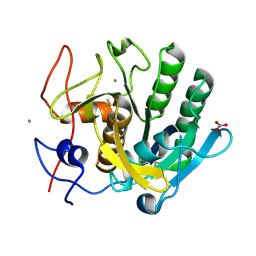 | | High-viscosity injector-based Pink Beam Serial Crystallography of Micro-crystals at a Synchrotron Radiation Source. | | Descriptor: | CALCIUM ION, NITRATE ION, Proteinase K | | Authors: | Martin-Garcia, J.M, Zhu, L, Mendez, D, Lee, M, Chun, E, Li, C, Hu, H, Subramanian, G, Kissick, D, Ogata, C, Henning, R, Ishchenko, A, Dobson, Z, Zhan, S, Weierstall, U, Spence, J.C.H, Fromme, P, Zatsepin, N.A, Fischetti, R.F, Cherezov, V, Liu, W. | | Deposit date: | 2018-09-17 | | Release date: | 2019-04-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | High-viscosity injector-based pink-beam serial crystallography of microcrystals at a synchrotron radiation source.
Iucrj, 6, 2019
|
|
7RSB
 
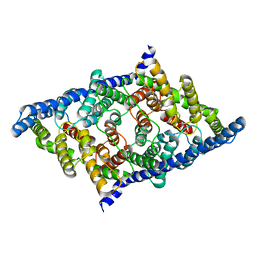 | |
4ZLW
 
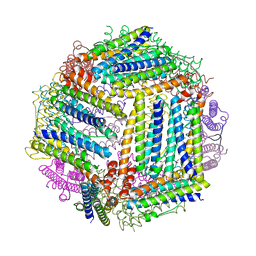 | |
6MHF
 
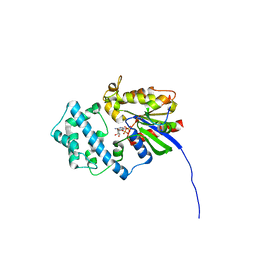 | | Galphai3 co-crystallized with GIV/Girdin | | Descriptor: | GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, Girdin, ... | | Authors: | Rees, S.D, Kalogriopoulos, N.A, Ngo, T, Kopcho, N, Ilatovskiy, A, Sun, N, Komives, E, Chang, G, Ghosh, P, Kufareva, I. | | Deposit date: | 2018-09-17 | | Release date: | 2019-07-31 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for GPCR-independent activation of heterotrimeric Gi proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5JE6
 
 | | Crystal structure of Burkholderia glumae ToxA | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Methyl transferase | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-11 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.571 Å) | | Cite: | Burkholderia glumae ToxA Is a Dual-Specificity Methyltransferase That Catalyzes the Last Two Steps of Toxoflavin Biosynthesis.
Biochemistry, 55, 2016
|
|
5N6W
 
 | | Retinoschisin R141H Mutant | | Descriptor: | Retinoschisin | | Authors: | Ramsay, E.P, Collins, R.F, Owens, T.W, Siebert, C.A, Jones, R.P.O, Roseman, A, Wang, T, Baldock, C. | | Deposit date: | 2017-02-16 | | Release date: | 2017-04-12 | | Last modified: | 2017-08-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of X-linked retinoschisis mutations reveals distinct classes which differentially effect retinoschisin function
Human Molecular Genetics, 25, 2016
|
|
