7O70
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(4R,7S)-12-chloro-14-fluoro-13-(2-fluoro-6-hydroxyphenyl)-4-methyl-10-oxa-2,5,16,18-tetrazatetracyclo[9.7.1.0^(2,7).0^(15,19)]nonadeca-1(18),11,13,15(19),16-pentaen-5-en-1-one-yl]prop-2, CALCIUM ION, GTPase KRas, ... | | Authors: | Phillips, C. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Discovery of AZD4625, a Covalent Allosteric Inhibitor of the Mutant GTPase KRAS G12C .
J.Med.Chem., 65, 2022
|
|
6UPP
 
 | | Radiation Damage Test of PixJ Pb state crystals | | Descriptor: | 1,2-ETHANEDIOL, MAGNESIUM ION, Methyl-accepting chemotaxis protein, ... | | Authors: | Clinger, J.A, Miller, M.D, Burgie, E.S, Vierstra, R.D, Phillips Jr, G.N. | | Deposit date: | 2019-10-18 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Photoreversible interconversion of a phytochrome photosensory module in the crystalline state.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7SX1
 
 | | human triosephosphate isomerase mutant v154m | | Descriptor: | ISOPROPYL ALCOHOL, Triosephosphate isomerase | | Authors: | Romero, J.M. | | Deposit date: | 2021-11-22 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | human triosephosphate isomerase mutant v154m
To Be Published
|
|
6PAN
 
 | |
8Q6G
 
 | | HUMAN PI4KIIIB IN COMPLEX WITH COVALENTLY BOUND INHIBITOR (COMPOUND 8) | | Descriptor: | 1,2-ETHANEDIOL, 3-(3,4-dimethoxyphenyl)-7-[(4-fluorosulfonyloxyphenyl)methylamino]-2,5-dimethyl-pyrazolo[1,5-a]pyrimidine, MAGNESIUM ION, ... | | Authors: | Somers, D.O. | | Deposit date: | 2023-08-11 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.541 Å) | | Cite: | Covalent targeting of non-cysteine residues in PI4KIII beta.
Rsc Chem Biol, 4, 2023
|
|
5C4R
 
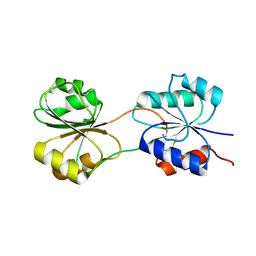 | | CobK precorrin-6A reductase | | Descriptor: | Precorrin-6A reductase | | Authors: | Gu, S, Pickersgill, R.W. | | Deposit date: | 2015-06-18 | | Release date: | 2016-11-16 | | Last modified: | 2017-08-30 | | Method: | X-RAY DIFFRACTION (3.17 Å) | | Cite: | Crystal structure of CobK reveals strand-swapping between Rossmann-fold domains and molecular basis of the reduced precorrin product trap.
Sci Rep, 5, 2015
|
|
7OCQ
 
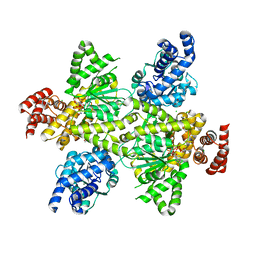 | | NADH bound to the dehydrogenase domain of the bifunctional mannitol-1-phosphate dehydrogenase/phosphatase MtlD from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tam, H.K, Mueller, V, Pos, K.M. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unidirectional mannitol synthesis of Acinetobacter baumannii MtlD is facilitated by the helix-loop-helix-mediated dimer formation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6UQ1
 
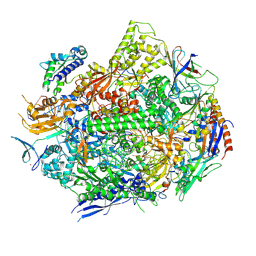 | | RNA polymerase II elongation complex with 5-guanidinohydantoin lesion in state 6 | | Descriptor: | DNA-directed RNA polymerase II subunit RPB1, DNA-directed RNA polymerase II subunit RPB11, DNA-directed RNA polymerase II subunit RPB2, ... | | Authors: | Oh, J, Wang, D. | | Deposit date: | 2019-10-18 | | Release date: | 2020-06-10 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6MED
 
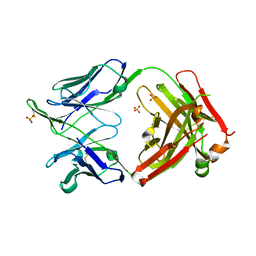 | | Crystal structure of broadly neutralizing antibody HEPC3 | | Descriptor: | SULFATE ION, antibody HEPC3 Heavy Chain, antibody HEPC3 Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J. | | Deposit date: | 2018-09-06 | | Release date: | 2018-11-21 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | HCV Broadly Neutralizing Antibodies Use a CDRH3 Disulfide Motif to Recognize an E2 Glycoprotein Site that Can Be Targeted for Vaccine Design.
Cell Host Microbe, 24, 2018
|
|
8Q6H
 
 | | HUMAN PI4KIIIB IN COMPLEX WITH COVALENTLY BOUND INHIBITOR (COMPOUND 11) | | Descriptor: | 1,2-ETHANEDIOL, 3-(3-fluorosulfonyloxy-4-methoxy-phenyl)-7-[(4-fluorosulfonyloxyphenyl)methylamino]-2,5-dimethyl-pyrazolo[1,5-a]pyrimidine, MAGNESIUM ION, ... | | Authors: | Somers, D.O. | | Deposit date: | 2023-08-11 | | Release date: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Covalent targeting of non-cysteine residues in PI4KIII beta.
Rsc Chem Biol, 4, 2023
|
|
6MEI
 
 | |
6PBB
 
 | | Crystal structure of Hen Egg White Lysozyme in complex with I3C | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CHLORIDE ION, Lysozyme | | Authors: | Truong, J.Q. | | Deposit date: | 2019-06-13 | | Release date: | 2019-07-10 | | Last modified: | 2020-01-01 | | Method: | X-RAY DIFFRACTION (1.89151084 Å) | | Cite: | Combining random microseed matrix screening and the magic triangle for the efficient structure solution of a potential lysin from bacteriophage P68.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
6MEN
 
 | | Crystal structure of a Tylonycteris bat coronavirus HKU4 macrodomain in complex with adenosine diphosphate glucose (ADP-glucose) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, Replicase polyprotein 1ab | | Authors: | Hammond, R.G, Schormann, N, McPherson, R.L, Leung, A.K.L, Deivanayagam, C.C.S, Johnson, M.A. | | Deposit date: | 2018-09-06 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | ADP-Ribose and Analogues bound to the DeMARylating Macrodomain from the Bat Coronavirus HKU4
Proc.Natl.Acad.Sci.USA, 2021
|
|
6USI
 
 | | Barrier-to-autointegration factor soaked in 1,6-hexanediol: 1 of 14 in MSCS set | | Descriptor: | Barrier-to-autointegration factor, ETHANOL | | Authors: | Agarwal, S, Smith, M, De La Rosa, I, Kliment, A.V, Swartz, P, Segura-Totten, M, Mattos, C. | | Deposit date: | 2019-10-26 | | Release date: | 2020-10-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.653 Å) | | Cite: | Development of a structure-analysis pipeline using multiple-solvent crystal structures of barrier-to-autointegration factor.
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6AUW
 
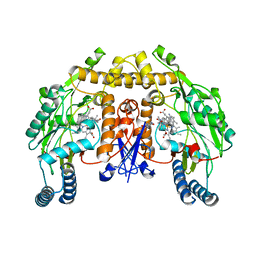 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 4-Methyl-6-(2-(5-(4-((methylamino)methyl)phenyl)pyridin-3-yl)ethyl)pyridin-2-amine | | Descriptor: | 4-methyl-6-[2-(5-{4-[(methylamino)methyl]phenyl}pyridin-3-yl)ethyl]pyridin-2-amine, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
7OCN
 
 | | Crystal structure of the bifunctional mannitol-1-phosphate dehydrogenase/phosphatase MtlD from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, BROMIDE ION, ... | | Authors: | Tam, H.K, Mueller, V, Pos, K.M. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unidirectional mannitol synthesis of Acinetobacter baumannii MtlD is facilitated by the helix-loop-helix-mediated dimer formation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
7OCR
 
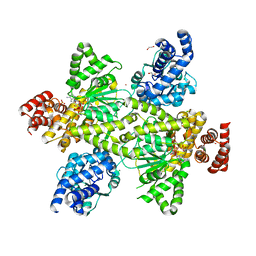 | | NADPH and fructose-6-phosphate bound to the dehydrogenase domain of the bifunctional mannitol-1-phosphate dehydrogenase/phosphatase MtlD from Acinetobacter baumannii | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Tam, H.K, Mueller, V, Pos, K.M. | | Deposit date: | 2021-04-28 | | Release date: | 2022-04-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Unidirectional mannitol synthesis of Acinetobacter baumannii MtlD is facilitated by the helix-loop-helix-mediated dimer formation.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
6UVX
 
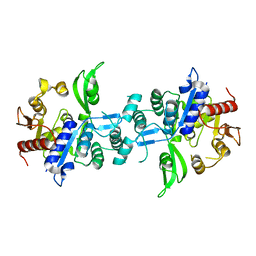 | | The crystal structure of FbiA from Mycobacterium Smegmatis, Apo state | | Descriptor: | CALCIUM ION, Phosphoenolpyruvate transferase | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6MFS
 
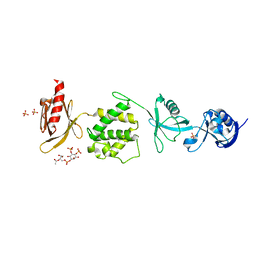 | | Mouse talin1 residues 1-138 fused to residues 169-400 in complex with phosphatidylinositol 4,5-bisphosphate (PIP2) | | Descriptor: | PHOSPHATE ION, Talin-1 fusion, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Izard, T, Chinthalapudi, K, Rangarajan, E.S. | | Deposit date: | 2018-09-12 | | Release date: | 2018-09-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The interaction of talin with the cell membrane is essential for integrin activation and focal adhesion formation.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8D6J
 
 | |
5C8V
 
 | |
7TEF
 
 | | Cytochrome P450 14 alpha-sterol demethylase CYP51 from Mycobacterium marinum | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Cytochrome P450 51B1 Cyp51B1, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Mohamed, H.A, Bruning, J.B, Bell, S.G. | | Deposit date: | 2022-01-04 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | A comparison of the bacterial CYP51 cytochrome P450 enzymes from Mycobacterium marinum and Mycobacterium tuberculosis.
J.Steroid Biochem.Mol.Biol., 221, 2022
|
|
6UW5
 
 | | The crystal structure of FbiA from Mycobacterium smegmatis, GDP and Fo bound form | | Descriptor: | 1-deoxy-1-(8-hydroxy-2,4-dioxo-3,4-dihydropyrimido[4,5-b]quinolin-10(2H)-yl)-D-ribitol, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
6MFV
 
 | | Crystal structure of the Signal Transduction ATPase with Numerous Domains (STAND) protein with a tetratricopeptide repeat sensor PH0952 from Pyrococcus horikoshii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, tetratricopeptide repeat sensor PH0952 | | Authors: | Lisa, M.N, Alzari, P.M, Haouz, A, Danot, O. | | Deposit date: | 2018-09-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Double autoinhibition mechanism of signal transduction ATPases with numerous domains (STAND) with a tetratricopeptide repeat sensor.
Nucleic Acids Res., 47, 2019
|
|
7SUB
 
 | | 3-oxoacyl-ACP reductase FabG | | Descriptor: | 3-oxoacyl-reductase, GLYCEROL | | Authors: | Thomas, L.M, Karr, E.A, Dinh, D.M. | | Deposit date: | 2021-11-16 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of a putative 3-hydroxypimelyl-CoA dehydrogenase, Hcd1, from Syntrophus aciditrophicus strain SB at 1.78 A resolution
Acta Crystallogr.,Sect.F, 79, 2023
|
|
