8P7D
 
 | | CryoEM structure of METTL6 tRNA SerRS complex in a 1:1:2 stoichiometry | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, Serine tRNA, ... | | Authors: | Throll, P, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8P7B
 
 | | CryoEM structure of METTL6 tRNA SerRS complex in a 1:2:2 stoichiometry | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, Serine tRNA, ... | | Authors: | Throll, P, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-12 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8P7C
 
 | | CryoEM structure of METTL6 tRNA SerRS complex in a 2:2:2 stoichiometry | | Descriptor: | MAGNESIUM ION, S-ADENOSYL-L-HOMOCYSTEINE, Serine tRNA, ... | | Authors: | Throll, P, Dolce, L.G, Kowalinski, E. | | Deposit date: | 2023-05-30 | | Release date: | 2024-06-12 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structural basis of tRNA recognition by the m 3 C RNA methyltransferase METTL6 in complex with SerRS seryl-tRNA synthetase.
Nat.Struct.Mol.Biol., 31, 2024
|
|
9I8V
 
 | | Cryo-EM structure of the Danio rerio tRNA ligase complex | | Descriptor: | ATP-dependent RNA helicase, Ashwin, Family with sequence similarity 98, ... | | Authors: | Chamera, S, Zajko, W, Czarnocki-Cieciura, M, Jaciuk, M, Koziej, L, Nowak, J, Wycisk, K, Sroka, M, Chramiec-Glabik, A, Smietanski, M, Golebiowski, F, Warminski, M, Jemielity, J, Glatt, S, Nowotny, M. | | Deposit date: | 2025-02-06 | | Release date: | 2025-04-23 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Structural and biochemical characterization of the 3'-5' tRNA splicing ligases.
J.Biol.Chem., 301, 2025
|
|
9DCH
 
 | | Single-stranded RNA-mediated PRC2 dimer | | Descriptor: | Isoform 2 of Histone-lysine N-methyltransferase EZH2, Polycomb protein EED, Polycomb protein SUZ12, ... | | Authors: | Song, J.S, Kasinath, V.K. | | Deposit date: | 2024-08-26 | | Release date: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Diverse RNA Structures Induce PRC2 Dimerization and Inhibit Histone Methyltransferase Activity.
Biorxiv, 2024
|
|
5FJ6
 
 | | Structure of the P2 polymerase inside in vitro assembled bacteriophage phi6 polymerase complex | | Descriptor: | MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Ilca, S, Kotecha, A, Sun, X, Poranen, M.P, Stuart, D.I, Huiskonen, J.T. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Localized Reconstruction of Subunits from Electron Cryomicroscopy Images of Macromolecular Complexes.
Nat.Commun., 6, 2015
|
|
8CWR
 
 | |
8CWT
 
 | |
1BT6
 
 | | P11 (S100A10), LIGAND OF ANNEXIN II IN COMPLEX WITH ANNEXIN II N-TERMINUS | | Descriptor: | ANNEXIN II, S100A10 | | Authors: | Rety, S, Sopkova, J, Renouard, M, Osterloh, D, Gerke, V, Russo-Marie, F, Lewit-Bentley, A. | | Deposit date: | 1998-09-02 | | Release date: | 1999-01-27 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of a complex of p11 with the annexin II N-terminal peptide.
Nat.Struct.Biol., 6, 1999
|
|
5FJ7
 
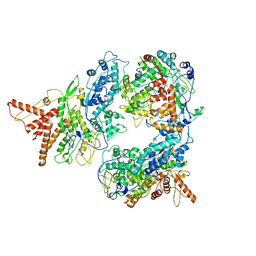 | | Structure of the P2 polymerase inside in vitro assembled bacteriophage phi6 polymerase complex, with P1 included | | Descriptor: | MAJOR INNER PROTEIN P1, MANGANESE (II) ION, RNA-DIRECTED RNA POLYMERASE | | Authors: | Ilca, S, Kotecha, A, Sun, X, Poranen, M.P, Stuart, D.I, Huiskonen, J.T. | | Deposit date: | 2015-10-06 | | Release date: | 2015-11-04 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (7.9 Å) | | Cite: | Localized Reconstruction of Subunits from Electron Cryomicroscopy Images of Macromolecular Complexes.
Nat.Commun., 6, 2015
|
|
5W1S
 
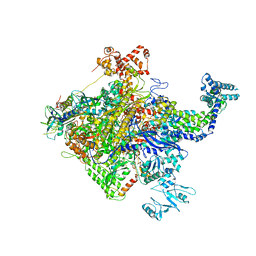 | |
6CUX
 
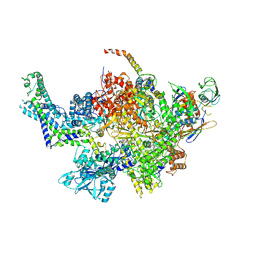 | |
3MF5
 
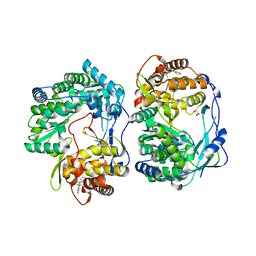 | |
5ZC9
 
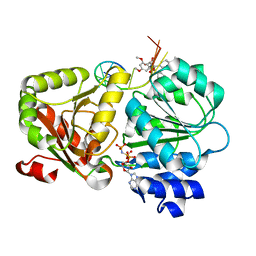 | | Crystal structure of the human eIF4A1-ATP analog-RocA-polypurine RNA complex | | Descriptor: | (1R,2R,3S,3aR,8bS)-6,8-dimethoxy-3a-(4-methoxyphenyl)-N,N-dimethyl-1,8b-bis(oxidanyl)-3-phenyl-2,3-dihydro-1H-cyclopenta[b][1]benzofuran-2-carboxamide, Eukaryotic initiation factor 4A-I, MAGNESIUM ION, ... | | Authors: | Iwasaki, W, Takahashi, M, Sakamoto, A, Iwasaki, S, Ito, T. | | Deposit date: | 2018-02-16 | | Release date: | 2019-01-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Translation Inhibitor Rocaglamide Targets a Bimolecular Cavity between eIF4A and Polypurine RNA.
Mol. Cell, 73, 2019
|
|
4AWK
 
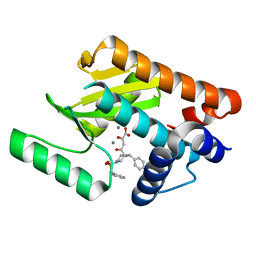 | | Influenza strain pH1N1 2009 polymerase subunit PA endonuclease in complex with diketo compound 1 | | Descriptor: | (2Z)-4-[(3S)-1-(benzylsulfonyl)-3-(4-chlorobenzyl)piperidin-3-yl]-2-hydroxy-4-oxobut-2-enoic acid, MANGANESE (II) ION, POLYMERASE PA | | Authors: | Kowalinski, E, Zubieta, C, Wolkerstorfer, A, Szolar, O.H, Ruigrok, R.W, Cusack, S. | | Deposit date: | 2012-06-04 | | Release date: | 2012-08-29 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of Specific Metal Chelating Inhibitor Binding to the Endonuclease Domain of Influenza Ph1N1 (2009) Polymerase.
Plos Pathog., 8, 2012
|
|
8KG8
 
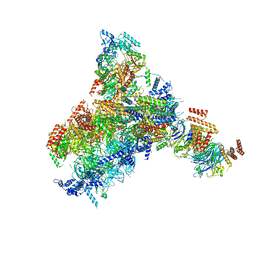 | | Yeast replisome in state II | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45, DNA (61-mer), ... | | Authors: | Dang, S, Zhai, Y, Feng, J, Yu, D, Xu, Z. | | Deposit date: | 2023-08-17 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (4.23 Å) | | Cite: | Synergism between CMG helicase and leading strand DNA polymerase at replication fork.
Nat Commun, 14, 2023
|
|
1Q79
 
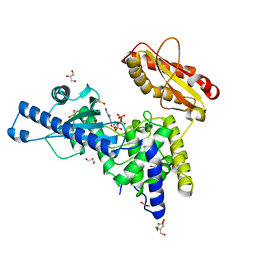 | | CRYSTAL STRUCTURE OF MAMMALIAN POLY(A) POLYMERASE | | Descriptor: | 3'-DEOXYADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Martin, G, Moglich, A, Keller, W, Doublie, S. | | Deposit date: | 2003-08-16 | | Release date: | 2004-09-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Biochemical and structural insights into substrate binding and catalytic mechanism of mammalian poly(A) polymerase.
J.Mol.Biol., 341, 2004
|
|
4AWM
 
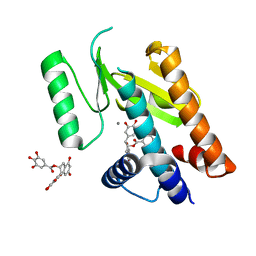 | | Influenza strain pH1N1 2009 polymerase subunit PA endonuclease in complex with (-)-epigallocatechin gallate from green tea | | Descriptor: | (2R,3R)-5,7-dihydroxy-2-(3,4,5-trihydroxyphenyl)-3,4-dihydro-2H-chromen-3-yl 3,4,5-trihydroxybenzoate, MANGANESE (II) ION, POLYMERASE PA | | Authors: | Kowalinski, E, Zubieta, C, Wolkerstorfer, A, Szolar, O.H, Ruigrok, R.W, Cusack, S. | | Deposit date: | 2012-06-04 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of Specific Metal Chelating Inhibitor Binding to the Endonuclease Domain of Influenza Ph1N1 (2009) Polymerase.
Plos Pathog., 8, 2012
|
|
3FBN
 
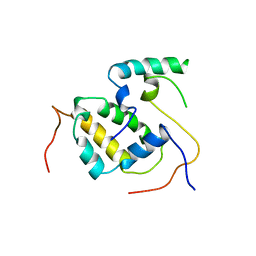 | | Structure of the Mediator submodule Med7N/31 | | Descriptor: | Mediator of RNA polymerase II transcription subunit 31, Mediator of RNA polymerase II transcription subunit 7 | | Authors: | Koschubs, T, Seizl, M, Lariviere, L, Kurth, F, Baumli, S, Martin, D.E, Cramer, P. | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.007 Å) | | Cite: | Identification, structure, and functional requirement of the Mediator submodule Med7N/31
Embo J., 28, 2009
|
|
4AWH
 
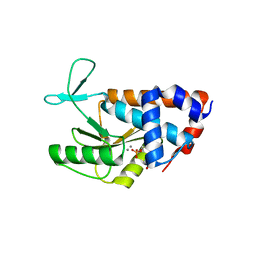 | | Influenza strain pH1N1 2009 polymerase subunit PA endonuclease in complex with rUMP | | Descriptor: | MANGANESE (II) ION, POLYMERASE PA, URIDINE-5'-MONOPHOSPHATE | | Authors: | Kowalinski, E, Zubieta, C, Wolkerstorfer, A, Szolar, O.H, Ruigrok, R.W, Cusack, S. | | Deposit date: | 2012-06-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Analysis of Specific Metal Chelating Inhibitor Binding to the Endonuclease Domain of Influenza Ph1N1 (2009) Polymerase.
Plos Pathog., 8, 2012
|
|
4AVG
 
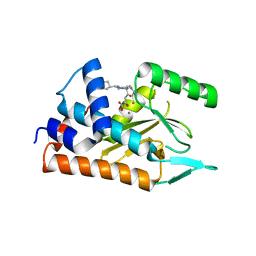 | | Influenza strain pH1N1 2009 polymerase subunit PA endonuclease in complex with diketo compound 2 | | Descriptor: | (Z)-4-[4-[(4-chlorophenyl)methyl]-1-(cyclohexylmethyl)piperidin-4-yl]-2-oxidanyl-4-oxidanylidene-but-2-enoic acid, MANGANESE (II) ION, POLYMERASE PA | | Authors: | Kowalinski, E, Zubieta, C, Wolkerstorfer, A, Szolar, O.H, Ruigrok, R.W, Cusack, S. | | Deposit date: | 2012-05-25 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Analysis of Specific Metal Chelating Inhibitor Binding to the Endonuclease Domain of Influenza Ph1N1 (2009) Polymerase.
Plos Pathog., 8, 2012
|
|
4AWF
 
 | | Influenza strain pH1N1 2009 polymerase subunit PA endonuclease in complex with 2 4-dioxo-4-phenylbutanoic acid DPBA | | Descriptor: | 2-4-DIOXO-4-PHENYLBUTANOIC ACID, MANGANESE (II) ION, POLYMERASE PA | | Authors: | Kowalinski, E, Zubieta, C, Wolkerstorfer, A, Szolar, O.H, Ruigrok, R.W, Cusack, S. | | Deposit date: | 2012-06-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Analysis of Specific Metal Chelating Inhibitor Binding to the Endonuclease Domain of Influenza Ph1N1 (2009) Polymerase.
Plos Pathog., 8, 2012
|
|
4AWG
 
 | | Influenza strain pH1N1 2009 polymerase subunit PA endonuclease in complex with diketo compound 3 | | Descriptor: | (2Z)-4-[(3S)-1-benzyl-3-(4-chlorobenzyl)piperidin-3-yl]-2-hydroxy-4-oxobut-2-enoic acid, MANGANESE (II) ION, POLYMERASE PA, ... | | Authors: | Kowalinski, E, Zubieta, C, Wolkerstorfer, A, Szolar, O.H, Ruigrok, R.W, Cusack, S. | | Deposit date: | 2012-06-03 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Analysis of Specific Metal Chelating Inhibitor Binding to the Endonuclease Domain of Influenza Ph1N1 (2009) Polymerase.
Plos Pathog., 8, 2012
|
|
4M5D
 
 | |
2BPC
 
 | | CRYSTAL STRUCTURE OF RAT DNA POLYMERASE BETA: EVIDENCE FOR A COMMON POLYMERASE MECHANISM | | Descriptor: | DNA POLYMERASE BETA, MANGANESE (II) ION | | Authors: | Sawaya, M.R, Pelletier, H, Kumar, A, Wilson, S.H, Kraut, J. | | Deposit date: | 1994-07-08 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of rat DNA polymerase beta: evidence for a common polymerase mechanism.
Science, 264, 1994
|
|
