7Y0P
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A/T268V/A82T/I263L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, p-cresol and hydroxylamine | | Descriptor: | (2S)-2-(6-imidazol-1-ylhexanoylamino)-3-phenyl-propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, HYDROXYAMINE, ... | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-06-06 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Crystal structure of the P450 BM3 heme domain mutant F87A/T268V/A82T/I263L in complex with N-imidazolyl-hexanoyl-L-phenylalanine, p-cresol and hydroxylamine
To Be Published
|
|
5TWX
 
 | | Crystal Structure of BRD9 bromodomain | | Descriptor: | Bromodomain-containing protein 9, N-[6-({2-[(3S)-2,6-dioxopiperidin-3-yl]-1,3-dioxo-2,3-dihydro-1H-isoindol-4-yl}oxy)hexyl]-2-(4-{2-[N-(1,1-dioxo-1lambda~6~-thian-4-yl)carbamimidoyl]-5-methyl-4-oxo-4,5-dihydrothieno[3,2-c]pyridin-7-yl}-2-methoxyphenoxy)acetamide | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2016-11-15 | | Release date: | 2017-09-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Degradation of the BAF Complex Factor BRD9 by Heterobifunctional Ligands.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
7P24
 
 | |
7XUW
 
 | | Crystal structure of RPA70N-BLMp2 fusion | | Descriptor: | fusion protein of Replication protein A 70 kDa DNA-binding subunit and Bloom syndrome protein | | Authors: | Wu, Y.Y, Zang, N, Fu, W.M, Zhou, C. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural characterization of human RPA70N association with DNA damage response proteins.
Elife, 12, 2023
|
|
8U6H
 
 | | Crystal Structure of HIV-1 Reverse Transcriptase in Complex with 3-(2-(2-(3-acryloyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-1-yl)ethoxy)-4-chlorophenoxy)-5-chlorobenzonitrile (JLJ744), a non-nucleoside inhibitor | | Descriptor: | 3-chloro-5-{4-chloro-2-[2-(2-oxo-3-propanoyl-2,3-dihydro-1H-benzimidazol-1-yl)ethoxy]phenoxy}benzonitrile, PHOSPHATE ION, Reverse transcriptase/ribonuclease H, ... | | Authors: | Prucha, G, Carter, Z, Jorgensen, W.L, Anderson, K.S. | | Deposit date: | 2023-09-13 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Covalent and noncovalent strategies for targeting Lys102 in HIV-1 reverse transcriptase.
Eur.J.Med.Chem., 262, 2023
|
|
5TXF
 
 | | Crystal structure of Lecithin:cholesterol acyltransferase (LCAT) in a closed conformation | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Phosphatidylcholine-sterol acyltransferase, ... | | Authors: | Manthei, K.A, Glukhova, A, Tesmer, J.J.G. | | Deposit date: | 2016-11-16 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A retractable lid in lecithin:cholesterol acyltransferase provides a structural mechanism for activation by apolipoprotein A-I.
J. Biol. Chem., 292, 2017
|
|
7Y0T
 
 | | Crystal structure of the P450 BM3 heme domain mutant F87A in complex with N-imidazolyl-hexanoyl-L-phenylalanyl-L-phenylalanine | | Descriptor: | Bifunctional cytochrome P450/NADPH--P450 reductase, I7X-PHE-PHE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Jiang, Y, Dong, S, Feng, Y, Cong, Z. | | Deposit date: | 2022-06-06 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Anchoring a Structurally Editable Proximal Cofactor-like Module to Construct an Artificial Dual-center Peroxygenase.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
7OZA
 
 | |
6MJH
 
 | |
8JV4
 
 | | Structure of the SAR11 PotD in complex with DMSP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(dimethyl-lambda~4~-sulfanyl)propanoic acid, GLYCEROL, ... | | Authors: | Ma, Q, Liu, C. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.445 Å) | | Cite: | Structure of the SAR11 PotD in complex with DMSP
To Be Published
|
|
7MTY
 
 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988569 | | Descriptor: | 2-(2,6-difluorophenyl)-5-hydroxy-N-[2-(2-methoxypyridin-4-yl)ethyl]-6-oxo-3,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-05-13 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
8JV1
 
 | | Structure of the SAR11 PotD in complex with GABA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Ma, Q, Liu, C. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.309 Å) | | Cite: | Structure of the SAR11 PotD in complex with GABA
To Be Published
|
|
6MHH
 
 | | Proteus mirabilis ScsC linker (residues 39-49) deletion and N6K mutant | | Descriptor: | Metal resistance protein | | Authors: | Furlong, E.J, Martin, J.L. | | Deposit date: | 2018-09-17 | | Release date: | 2019-03-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.083 Å) | | Cite: | Engineered variants provide new insight into the structural properties important for activity of the highly dynamic, trimeric protein disulfide isomerase ScsC from Proteus mirabilis.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
7OQ2
 
 | | NaK S-DI mutant soaked in Na+ | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Minniberger, S, Plested, A.J.R. | | Deposit date: | 2021-06-02 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Asymmetry and Ion Selectivity Properties of Bacterial Channel NaK Mutants Derived from Ionotropic Glutamate Receptors.
J.Mol.Biol., 435, 2023
|
|
7MU7
 
 | | Ask1 bound to compound 3 | | Descriptor: | 1,2-ETHANEDIOL, 3-methoxy-N-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}-1-(pyrazin-2-yl)-1H-pyrazole-4-carboxamide, DIMETHYL SULFOXIDE, ... | | Authors: | Chodaparambil, J.V, Marcotte, D.J. | | Deposit date: | 2021-05-14 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.298 Å) | | Cite: | Discovery of Potent, Selective and CNS-Penetrant Apoptosis Signal-regulating Kinase 1 (ASK1) Inhibitors that Modulate Brain Inflammation in Vivo
To Be Published
|
|
5UC7
 
 | | Crystal structure of BioA / 7,8-diaminopelargonic acid aminotransferase / DAPA synthase from Citrobacter rodentium, PLP complex | | Descriptor: | Adenosylmethionine-8-amino-7-oxononanoate aminotransferase, D(-)-TARTARIC ACID | | Authors: | Stogios, P.J, Evdokimova, E, Di Leo, R, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-12-21 | | Release date: | 2017-01-25 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.835 Å) | | Cite: | Crystal structure of BioA / 7,8-diaminopelargonic acid aminotransferase / DAPA synthase from Citrobacter rodentium, PLP complex
To Be Published
|
|
6MJP
 
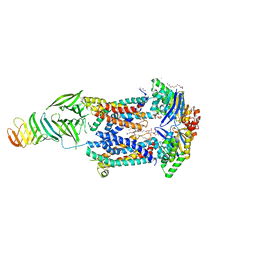 | | LptB(E163Q)FGC from Vibrio cholerae | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 6-cyclohexylhexyl beta-D-glucopyranoside, ABC transporter ATP-binding protein, ... | | Authors: | Owens, T.W, Kahne, D, Kruse, A.C. | | Deposit date: | 2018-09-21 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of unidirectional export of lipopolysaccharide to the cell surface.
Nature, 567, 2019
|
|
6MKF
 
 | | Crystal structure of penicillin binding protein 5 (PBP5) from Enterococcus faecium in the imipenem-bound form | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carbox ylic acid, SULFATE ION, penicillin binding protein 5 (PBP5) | | Authors: | Moon, T.M, Lee, C, D'Andrea, E.D, Peti, W, Page, R. | | Deposit date: | 2018-09-25 | | Release date: | 2018-10-31 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The structures of penicillin-binding protein 4 (PBP4) and PBP5 fromEnterococciprovide structural insights into beta-lactam resistance.
J. Biol. Chem., 293, 2018
|
|
7MS2
 
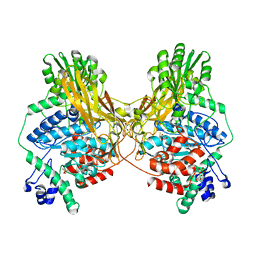 | |
5CNW
 
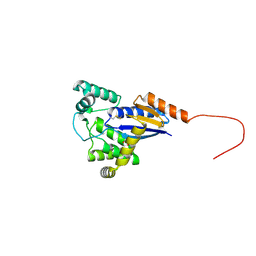 | |
7OQ8
 
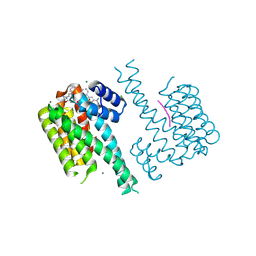 | |
8QPS
 
 | |
7OZE
 
 | |
5TYT
 
 | | Crystal Structure of the PDZ domain of RhoGEF bound to CXCR2 C-terminal peptide | | Descriptor: | Rho guanine nucleotide exchange factor 11, C-X-C chemokine receptor type 2 chimera | | Authors: | Spellmon, N, Holcomb, J, Niu, A, Choudhary, V, Sun, X, Brunzelle, J, Li, C, Yang, Z. | | Deposit date: | 2016-11-21 | | Release date: | 2017-02-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis of PDZ-mediated chemokine receptor CXCR2 scaffolding by guanine nucleotide exchange factor PDZ-RhoGEF.
Biochem. Biophys. Res. Commun., 485, 2017
|
|
8QOO
 
 | | Crystal structure of human Sirt2 in complex with the peptide-based pseudo-inhibitor TNFn-4.1 | | Descriptor: | (2S,3R)-3-dodecylsulfanyl-2-methyl-butanoic acid, (2S,3S)-3-dodecylsulfanyl-2-methyl-butanoic acid, (R,R)-2,3-BUTANEDIOL, ... | | Authors: | Friedrich, F, Kalbas, D, Meleshin, M, Einsle, O, Schutkowski, M, Jung, M. | | Deposit date: | 2023-09-29 | | Release date: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Development of potent TNFa-Myr-based pseudo-inhibitors for Sirtuin 2
To Be Published
|
|
