8XB3
 
 | |
6ZOK
 
 | | SARS-CoV-2-Nsp1-40S complex, focused on body | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S11, 40S ribosomal protein S13, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-29 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7V86
 
 | | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Kappa variant (B.1.617.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form
To Be Published
|
|
7V89
 
 | | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Angiotensin-converting enzyme 2 (ACE2) ectodomain, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Delta variant (B.1.617.2) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 1
To Be Published
|
|
7V82
 
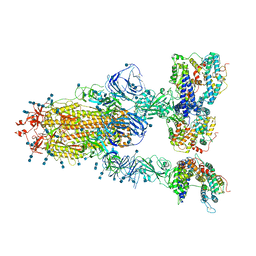 | | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 1
To Be Published
|
|
7V83
 
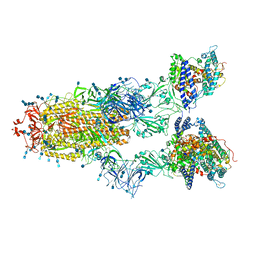 | | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2,Green fluorescent protein, ... | | Authors: | Yang, T.J, Yu, P.Y, Chang, Y.C, Hsu, S.T.D. | | Deposit date: | 2021-08-22 | | Release date: | 2021-10-06 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-EM structure of SARS-CoV-2 S-Gamma variant (P.1) in complex with Angiotensin-converting enzyme 2 (ACE2) ectodomain, three ACE2-bound form conformation 2
To Be Published
|
|
2ECK
 
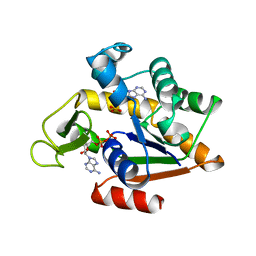 | | STRUCTURE OF PHOSPHOTRANSFERASE | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ADENYLATE KINASE | | Authors: | Berry, M.B, Bilderback, T, Glaser, M, Phillips Jr, G.N. | | Deposit date: | 1996-12-16 | | Release date: | 1997-03-12 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of ADP/AMP complex of Escherichia coli adenylate kinase.
Proteins, 62, 2006
|
|
5CGF
 
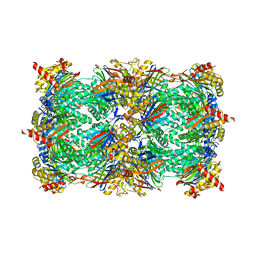 | | Yeast 20S proteasome beta5-G48C mutant | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Probable proteasome subunit alpha type-7, ... | | Authors: | Dubiella, C, Groll, M. | | Deposit date: | 2015-07-09 | | Release date: | 2015-11-25 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Selective Inhibition of the Immunoproteasome by Structure-Based Targeting of a Non-catalytic Cysteine.
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
6SGC
 
 | | Rabbit 80S ribosome stalled on a poly(A) tail | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S12, ... | | Authors: | Chandrasekaran, V, Juszkiewicz, S, Choi, J, Puglisi, J.D, Brown, A, Shao, S, Ramakrishnan, V, Hegde, R.S. | | Deposit date: | 2019-08-03 | | Release date: | 2019-12-04 | | Last modified: | 2022-03-30 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Mechanism of ribosome stalling during translation of a poly(A) tail.
Nat.Struct.Mol.Biol., 26, 2019
|
|
8G6J
 
 | | mRNA decoding in human is kinetically and structurally distinct from bacteria (GA state 2) | | Descriptor: | (3R,6R,9S,12S,15S,18S,20R,24aR)-6-[(2S)-butan-2-yl]-3,12-bis[(1R)-1-hydroxy-2-methylpropyl]-8,9,11,17,18-pentamethyl-15-[(2S)-2-methylbutyl]hexadecahydropyrido[1,2-a][1,4,7,10,13,16,19]heptaazacyclohenicosine-1,4,7,10,13,16,19(21H)-heptone, (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 1,4-DIAMINOBUTANE, ... | | Authors: | Holm, M, Natchiar, K.S, Rundlet, E.J, Myasnikov, A.G, Altman, R.B, Blanchard, S.C. | | Deposit date: | 2023-02-15 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | mRNA decoding in human is kinetically and structurally distinct from bacteria.
Nature, 617, 2023
|
|
6ZOJ
 
 | | SARS-CoV-2-Nsp1-40S complex, composite map | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S10, 40S ribosomal protein S11, ... | | Authors: | Schubert, K, Karousis, E.D, Jomaa, A, Scaiola, A, Echeverria, B, Gurzeler, L.-A, Leibundgut, M.L, Thiel, V, Muehlemann, O, Ban, N. | | Deposit date: | 2020-07-07 | | Release date: | 2020-07-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7UCK
 
 | | 80S translation initiation complex with ac4c(-1) mRNA and Harringtonine | | Descriptor: | 18S rRNA, 28s rRNA, 40S ribosomal protein S10, ... | | Authors: | Yang, R, Arango, D, Sturgill, D, Oberdoerffer, S. | | Deposit date: | 2022-03-16 | | Release date: | 2022-06-01 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Direct epitranscriptomic regulation of mammalian translation initiation through N4-acetylcytidine.
Mol.Cell, 82, 2022
|
|
9BDL
 
 | | 80S ribosome with angiogenin | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Korostelev, A.A. | | Deposit date: | 2024-04-12 | | Release date: | 2024-07-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural mechanism of angiogenin activation by the ribosome.
Nature, 630, 2024
|
|
6RMD
 
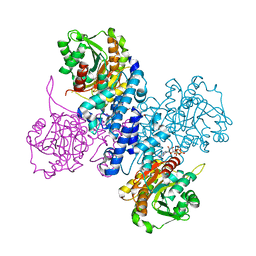 | | Structure of ATP bound Plasmodium falciparum IMP-nucleotidase | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-5'-TRIPHOSPHATE, IMP-specific 5'-nucleotidase, ... | | Authors: | Carrique, L, Ballut, L, Violot, S, Aghajari, N. | | Deposit date: | 2019-05-06 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and catalytic regulation of Plasmodium falciparum IMP specific nucleotidase.
Nat Commun, 11, 2020
|
|
7MJ2
 
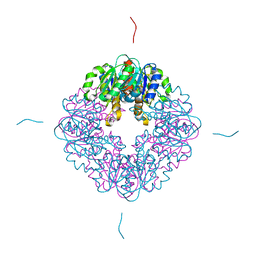 | | LarB, a carboxylase/hydrolase involved in synthesis of the cofactor for lactate racemase, in complex with Zn | | Descriptor: | MAGNESIUM ION, Pyridinium-3,5-biscarboxylic acid mononucleotide synthase, ZINC ION | | Authors: | Chatterjee, S, Rankin, J.A, Lagishetty, S, Hu, J, Hausinger, R.P. | | Deposit date: | 2021-04-19 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The LarB carboxylase/hydrolase forms a transient cysteinyl-pyridine intermediate during nickel-pincer nucleotide cofactor biosynthesis.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4JRV
 
 | | Crystal structure of EGFR kinase domain in complex with compound 4c | | Descriptor: | 4-(dimethylamino)-N-[3-(4-{[(1S)-2-hydroxy-1-phenylethyl]amino}-6-phenylfuro[2,3-d]pyrimidin-5-yl)phenyl]butanamide, Epidermal growth factor receptor | | Authors: | Peng, Y.H, Wu, J.S. | | Deposit date: | 2013-03-22 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Protein Kinase Inhibitor Design by Targeting the Asp-Phe-Gly (DFG) Motif: The Role of the DFG Motif in the Design of Epidermal Growth Factor Receptor Inhibitors
J.Med.Chem., 56, 2013
|
|
6SNT
 
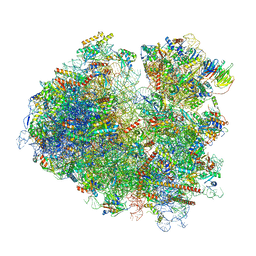 | | Yeast 80S ribosome stalled on SDD1 mRNA. | | Descriptor: | 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, 40S ribosomal protein S10-A, ... | | Authors: | Tesina, P, Buschauer, R, Cheng, J, Becker, T, Beckmann, R. | | Deposit date: | 2019-08-27 | | Release date: | 2020-03-04 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | RQT complex dissociates ribosomes collided on endogenous RQC substrate SDD1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7FHO
 
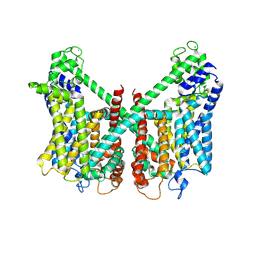 | | Structure of AtTPC1 D240A/D454A/E528A mutant with 50 mM Ca2+ | | Descriptor: | CALCIUM ION, Two pore calcium channel protein 1,GFP | | Authors: | Ye, F, Xu, L, Li, X, Jiang, Y, Guo, J. | | Deposit date: | 2021-07-29 | | Release date: | 2021-12-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Voltage-gating and cytosolic Ca 2+ activation mechanisms of Arabidopsis two-pore channel AtTPC1.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
4Z4K
 
 | |
4TS0
 
 | | Crystal structure of the Spinach RNA aptamer in complex with DFHBI, barium ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2,3-dimethyl-3,5-dihydro-4H-imidazol-4-one, BARIUM ION, POTASSIUM ION, ... | | Authors: | Warner, K.D, Chen, M.C, Song, W, Strack, R.L, Thorn, A, Jaffrey, S.R, Ferre-D'Amare, A.R. | | Deposit date: | 2014-06-18 | | Release date: | 2014-07-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for activity of highly efficient RNA mimics of green fluorescent protein.
Nat.Struct.Mol.Biol., 21, 2014
|
|
6DGV
 
 | |
3X2S
 
 | | Crystal structure of pyrene-conjugated adenylate kinase | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Fujii, A, Sekiguchi, Y, Matsumura, H, Inoue, T, Chung, W.-S, Hirota, S, Matsuo, T. | | Deposit date: | 2014-12-31 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Excimer Emission Properties on Pyrene-Labeled Protein Surface: Correlation between Emission Spectra, Ring Stacking Modes, and Flexibilities of Pyrene Probes.
Bioconjug.Chem., 26, 2015
|
|
2JEE
 
 | | Xray structure of E. coli YiiU | | Descriptor: | CELL DIVISION PROTEIN ZAPB | | Authors: | Moller-Jensen, J, Gerdes, K, Lowe, J. | | Deposit date: | 2007-01-16 | | Release date: | 2008-02-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel Coiled-Coil Cell Division Factor Zapb Stimulates Z Ring Assembly and Cell Division.
Mol.Microbiol., 68, 2008
|
|
3OGO
 
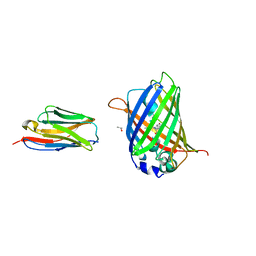 | | Structure of the GFP:GFP-nanobody complex at 2.8 A resolution in spacegroup P21212 | | Descriptor: | GFP-nanobody, Green fluorescent protein, ISOPROPYL ALCOHOL | | Authors: | Kubala, M.H, Kovtun, O, Alexandrov, K, Collins, B.M. | | Deposit date: | 2010-08-17 | | Release date: | 2010-08-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural and thermodynamic analysis of the GFP:GFP-nanobody complex.
Protein Sci., 19, 2010
|
|
4BD7
 
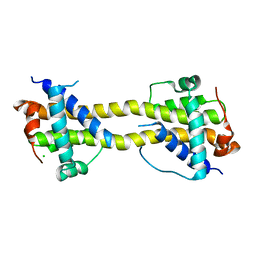 | | Bax domain swapped dimer induced by octylmaltoside | | Descriptor: | APOPTOSIS REGULATOR BAX, CHLORIDE ION, PRASEODYMIUM ION | | Authors: | Czabotar, P.E, Westphal, D, Adams, J.M, Colman, P.M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Bax Crystal Structures Reveal How Bh3 Domains Activate Bax and Nucleate its Oligomerization to Induce Apoptosis.
Cell(Cambridge,Mass.), 152, 2013
|
|
