1Z2V
 
 | |
1AFC
 
 | | STRUCTURAL STUDIES OF THE BINDING OF THE ANTI-ULCER DRUG SUCROSE OCTASULFATE TO ACIDIC FIBROBLAST GROWTH FACTOR | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, ACIDIC FIBROBLAST GROWTH FACTOR | | Authors: | Zhu, X, Hsu, B.T, Rees, D.C. | | Deposit date: | 1993-07-13 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural studies of the binding of the anti-ulcer drug sucrose octasulfate to acidic fibroblast growth factor.
Structure, 1, 1993
|
|
1NOU
 
 | | Native human lysosomal beta-hexosaminidase isoform B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, SULFATE ION, ... | | Authors: | Mark, B.L, Mahuran, D.J, Cherney, M.M, Zhao, D, Knapp, S, James, M.N.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-04-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of Human beta-hexosaminidase B: Understanding the molecular basis of Sandhoff and Tay-Sachs disease
J.Mol.Biol., 327, 2003
|
|
1OCP
 
 | | SOLUTION STRUCTURE OF OCT3 POU-HOMEODOMAIN | | Descriptor: | OCT-3 | | Authors: | Morita, E.H, Hayashi, F, Shirakawa, M, Kyogoku, Y. | | Deposit date: | 1995-02-21 | | Release date: | 1995-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the Oct-3 POU-Homeodomain in Solution, as Determined by Triple Resonance Heteronuclear Multidimensional NMR Spectroscopy
Protein Sci., 4, 1995
|
|
1NZ8
 
 | | Solution Structure of the N-utilization substance G (NusG) N-terminal (NGN) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
1OCT
 
 | | CRYSTAL STRUCTURE OF THE OCT-1 POU DOMAIN BOUND TO AN OCTAMER SITE: DNA RECOGNITION WITH TETHERED DNA-BINDING MODULES | | Descriptor: | DNA (5'-D(*AP*CP*CP*TP*TP*AP*TP*TP*TP*GP*CP*AP*TP*AP*C)-3'), DNA (5'-D(*TP*GP*TP*AP*TP*GP*CP*AP*AP*AP*TP*AP*AP*GP*G)-3'), PROTEIN (OCT-1 POU DOMAIN) | | Authors: | Klemm, J.D, Rould, M.A, Aurora, R, Herr, W, Pabo, C.O. | | Deposit date: | 1994-05-09 | | Release date: | 1994-08-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA-binding modules.
Cell(Cambridge,Mass.), 77, 1994
|
|
1Q41
 
 | | GSK-3 Beta complexed with Indirubin-3'-monoxime | | Descriptor: | (Z)-1H,1'H-[2,3']BIINDOLYLIDENE-3,2'-DIONE-3-OXIME, GLYCOGEN SYNTHASE KINASE-3 BETA | | Authors: | Bertrand, J.A, Thieffine, S, Vulpetti, A, Cristiani, C, Valsasina, B, Knapp, S, Kalisz, H.M, Flocco, M. | | Deposit date: | 2003-08-01 | | Release date: | 2003-10-21 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural Characterization of the Gsk-3Beta Active Site Using Selective and Non-selective ATP-Mimetic Inhibitors
J.Mol.Biol., 333, 2003
|
|
1YKR
 
 | | Crystal structure of cdk2 with an aminoimidazo pyridine inhibitor | | Descriptor: | 4-{[6-(2,6-DICHLOROBENZOYL)IMIDAZO[1,2-A]PYRIDIN-2-YL]AMINO}BENZENESULFONAMIDE, Cell division protein kinase 2 | | Authors: | Hamdouchi, C, Zhong, B, Mendoza, J, Jaramillo, C, Zhang, F, Brooks, H.B. | | Deposit date: | 2005-01-18 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure-based design of a new class of highly selective aminoimidazo[1,2-a]pyridine-based inhibitors of cyclin dependent kinases
Bioorg.Med.Chem.Lett., 15, 2005
|
|
1PYC
 
 | | CYP1 (HAP1) DNA-BINDING DOMAIN (RESIDUES 60-100), NMR, 15 STRUCTURES | | Descriptor: | CYP1, ZINC ION | | Authors: | Timmerman, J, Vuidepot, A.-L, Bontems, F, Lallemand, J.-Y, Gervais, M, Shechter, E, Guiard, B. | | Deposit date: | 1996-02-17 | | Release date: | 1996-08-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H, 15N resonance assignment and three-dimensional structure of CYP1 (HAP1) DNA-binding domain.
J.Mol.Biol., 259, 1996
|
|
1POG
 
 | | SOLUTION STRUCTURE OF THE OCT-1 POU-HOMEO DOMAIN DETERMINED BY NMR AND RESTRAINED MOLECULAR DYNAMICS | | Descriptor: | OCT-1 POU HOMEODOMAIN DNA-BINDING PROTEIN | | Authors: | Cox, M, Van Tilborg, P.J.A, De Laat, W, Boelens, R, Van Leeuwen, H.C, Van Der Vliet, P.C, Kaptein, R. | | Deposit date: | 1994-10-12 | | Release date: | 1995-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Oct-1 POU homeodomain determined by NMR and restrained molecular dynamics.
J.Biomol.NMR, 6, 1995
|
|
1YO5
 
 | | Analysis of the 2.0A crystal structure of the protein-DNA complex of human PDEF Ets domain bound to the prostate specific antigen regulatory site | | Descriptor: | Enhancer site of Prostate Specific Antigen Promoter Region, SAM pointed domain containing ets transcription factor | | Authors: | Wang, Y, Feng, L, Said, M, Balderman, S, Fayazi, Z, Liu, Y, Ghosh, D, Gulick, A.M. | | Deposit date: | 2005-01-26 | | Release date: | 2005-05-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Analysis of the 2.0 A Crystal Structure of the Protein-DNA Complex of the Human PDEF Ets Domain Bound to the Prostate Specific Antigen Regulatory Site
Biochemistry, 44, 2005
|
|
1OL1
 
 | | Cyclin A binding groove inhibitor H-Cit-Cit-Leu-Ile-(p-F-Phe)-NH2 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CIR-CIR-LEU-ILE-PFF-NH2, CYCLIN A2 | | Authors: | Kontopidis, G, Andrews, M, McInnes, C, Cowan, A, Powers, H, Innes, L, Plater, A, Griffiths, G, Paterson, D, Zheleva, D, Lane, D, Green, S, Walkinshaw, M, Fischer, P. | | Deposit date: | 2003-08-04 | | Release date: | 2003-12-11 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insights Into Cyclin Groove Recognition. Complex Crystal Structures and Inhibitor Design Through Ligand Exchange
Structure, 11, 2003
|
|
1OIY
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | Descriptor: | 4-(6-CYCLOHEXYLMETHOXY-9H-PURIN-2-YLAMINO)--BENZAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-06-26 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
1OI9
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | Descriptor: | 6-CYCLOHEXYLMETHYLOXY-2-(4'-HYDROXYANILINO)PURINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-06-10 | | Release date: | 2004-07-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
1OIU
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with a 6-cyclohexylmethyloxy-2-anilino-purine inhibitor | | Descriptor: | 3-(6-CYCLOHEXYLMETHOXY-9H-PURIN-2-YLAMINO)-BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-06-26 | | Release date: | 2004-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | N2-Substituted O6-Cyclohexylmethylguanine Derivatives: Potent Inhibitors of Cyclin-Dependent Kinases 1 and 2
J.Med.Chem., 47, 2004
|
|
1OKV
 
 | | Cyclin A binding groove inhibitor H-Arg-Arg-Leu-Ile-Phe-NH2 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, H-ARG-ARG-LEU-ILE-PHE-NH2 | | Authors: | Kontopidis, G, Andrews, M, McInnes, C, Cowan, A, Powers, H, Innes, L, Plater, A, Griffiths, G, Paterson, D, Zheleva, D, Lane, D, Green, S, Walkinshaw, M, Fischer, P. | | Deposit date: | 2003-07-30 | | Release date: | 2003-12-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Insights Into Cyclin Groove Recognition. Complex Crystal Structures and Inhibitor Design Through Ligand Exchange
Structure, 11, 2003
|
|
1OGU
 
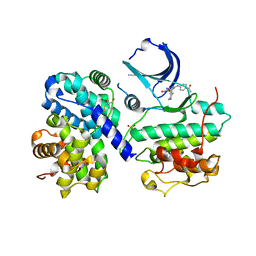 | | STRUCTURE OF HUMAN THR160-PHOSPHO CDK2/CYCLIN A COMPLEXED WITH A 2-ARYLAMINO-4-CYCLOHEXYLMETHYL-5-NITROSO-6-AMINOPYRIMIDINE INHIBITOR | | Descriptor: | 4-{[4-AMINO-6-(CYCLOHEXYLMETHOXY)-5-NITROSOPYRIMIDIN-2-YL]AMINO}BENZAMIDE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, ... | | Authors: | Pratt, D.J, Endicott, J.A, Noble, M.E.M. | | Deposit date: | 2003-05-13 | | Release date: | 2003-09-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structure-Based Design of 2-Arylamino-4-Cyclohexyl Methyl-5-Nitroso-6-Aminopyrimidine Inhibitors of Cyclin-Dependent Kinases 1 and 2
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1OIR
 
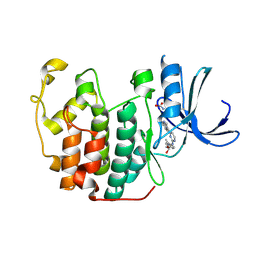 | | Imidazopyridines: a potent and selective class of Cyclin-dependent Kinase inhibitors identified through Structure-based hybridisation | | Descriptor: | 1-(DIMETHYLAMINO)-3-(4-{{4-(2-METHYLIMIDAZO[1,2-A]PYRIDIN-3-YL)PYRIMIDIN-2-YL]AMINO}PHENOXY)PROPAN-2-OL, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Beattie, J.F, Breault, G.A, Byth, K.F, Culshaw, J.D, Ellston, R.P.A, Green, S, Minshull, C.A, Norman, R.A, Pauptit, R.A, Thomas, A.P, Jewsbury, P.J. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Imidazo[1,2-A]Pyridines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors Identified Through Structure-Based Hybridisation
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1NOW
 
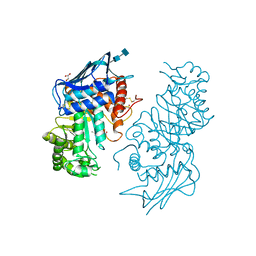 | | Human lysosomal beta-hexosaminidase isoform B in complex with (2R,3R,4S,5R)-2-Acetamido-3,4-Dihydroxy-5-Hydroxymethyl-Piperidinium Chloride (GalNAc-isofagomine) | | Descriptor: | (2R,3R,4S,5R)-2-ACETAMIDO-3,4-DIHYDROXY-5-HYDROXYMETHYL-PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Mark, B.L, Mahuran, D.J, Cherney, M.M, Zhao, D, Knapp, S, James, M.N.G. | | Deposit date: | 2003-01-16 | | Release date: | 2003-04-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Human beta-hexosaminidase B: Understanding the molecular basis of Sandhoff and Tay-Sachs disease
J.Mol.Biol., 327, 2003
|
|
1OIT
 
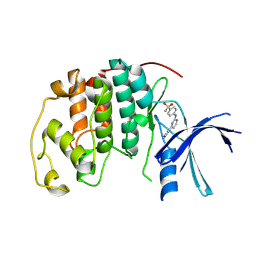 | | Imidazopyridines: a potent and selective class of Cyclin-dependent Kinase inhibitors identified through Structure-based hybridisation | | Descriptor: | 4-[(4-IMIDAZO[1,2-A]PYRIDIN-3-YLPYRIMIDIN-2-YL)AMINO]BENZENESULFONAMIDE, CELL DIVISION PROTEIN KINASE 2 | | Authors: | Beattie, J.F, Breault, G.A, Byth, K.F, Culshaw, J.D, Ellston, R.P.A, Green, S, Minshull, C.A, Norman, R.A, Pauptit, R.A, Thomas, A.P, Jewsbury, P.J. | | Deposit date: | 2003-06-24 | | Release date: | 2003-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Imidazo[1,2-A]Pyridines: A Potent and Selective Class of Cyclin-Dependent Kinase Inhibitors Identified Through Structure-Based Hybridisation
Bioorg.Med.Chem.Lett., 13, 2003
|
|
1ZTR
 
 | | Solution structure of Engrailed homeodomain L16A mutant | | Descriptor: | Segmentation polarity homeobox protein engrailed | | Authors: | Religa, T.L, Markson, J.S, Mayor, U, Freund, S.M.V, Fersht, A.R. | | Deposit date: | 2005-05-27 | | Release date: | 2005-10-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a protein denatured state and folding intermediate.
Nature, 437, 2005
|
|
1NZ9
 
 | | Solution Structure of the N-utilization substance G (NusG) C-terminal (NGC) domain from Thermus thermophilus | | Descriptor: | TRANSCRIPTION ANTITERMINATION PROTEIN NUSG | | Authors: | Reay, P, Yamasaki, K, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-02-17 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and sequence comparisons arising from the solution structure of the transcription elongation factor NusG from Thermus thermophilus
Proteins, 56, 2004
|
|
1O9K
 
 | | Crystal structure of the retinoblastoma tumour suppressor protein bound to E2F peptide | | Descriptor: | RETINOBLASTOMA-ASSOCIATED PROTEIN, TRANSCRIPTION FACTOR E2F1 | | Authors: | Xiao, B, Spencer, J, Clements, A, Ali-Khan, N, Mittnacht, S, Broceno, C, Burghammer, M, Perrakis, A, Marmorstein, R, Gamblin, S.J. | | Deposit date: | 2002-12-16 | | Release date: | 2003-03-06 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Retinoblastoma Tumor Suppressor Protein Bound to E2F and the Molecular Basis of its Regulation
Proc.Natl.Acad.Sci.USA, 100, 2003
|
|
1YAZ
 
 | | AZIDE-BOUND YEAST CU(II)/ZN SUPEROXIDE DISMUTASE ROOM TEMPERATURE (298K) STRUCTURE | | Descriptor: | AZIDE ION, COPPER (II) ION, PROTEIN (CU/ZN SUPEROXIDE DISMUTASE), ... | | Authors: | Hart, P.J, Balbirnie, M.M, Ogihara, N.L, Nersissian, A.M, Weiss, M.S, Valentine, J.S, Eisenberg, D. | | Deposit date: | 1998-12-23 | | Release date: | 2000-01-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A structure-based mechanism for copper-zinc superoxide dismutase.
Biochemistry, 38, 1999
|
|
1OM9
 
 | | Structure of the GGA1-appendage in complex with the p56 binding peptide | | Descriptor: | 15-mer peptide fragment of p56, ADP-ribosylation factor binding protein GGA1 | | Authors: | Collins, B.M, Praefcke, G.J.K, Robinson, M.S, Owen, D.J. | | Deposit date: | 2003-02-25 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for binding of accessory proteins by the appendage domain of GGAs
Nat.Struct.Biol., 10, 2003
|
|
