3E91
 
 | |
3E2M
 
 | | LFA-1 I domain bound to inhibitors | | Descriptor: | Integrin alpha-L, cis-4-{[2-({4-[(1E)-3-morpholin-4-yl-3-oxoprop-1-en-1-yl]-2,3-bis(trifluoromethyl)phenyl}sulfanyl)phenoxy]methyl}cyclohexanecarboxylic acid | | Authors: | Silvian, L.F. | | Deposit date: | 2008-08-05 | | Release date: | 2008-08-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-activity relationship of ortho- and meta-phenol based LFA-1 ICAM inhibitors
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3E2S
 
 | | Crystal Structure Reduced PutA86-630 Mutant Y540S Complexed with L-proline | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, PENTAETHYLENE GLYCOL, PROLINE, ... | | Authors: | Tanner, J.J. | | Deposit date: | 2008-08-06 | | Release date: | 2009-02-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A conserved active site tyrosine residue of proline dehydrogenase helps enforce the preference for proline over hydroxyproline as the substrate.
Biochemistry, 48, 2009
|
|
3E3A
 
 | | The Structure of Rv0554 from Mycobacterium tuberculosis | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, POSSIBLE PEROXIDASE BPOC | | Authors: | Johnston, J.M, Baker, E.N. | | Deposit date: | 2008-08-06 | | Release date: | 2009-06-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural and functional analysis of Rv0554 from Mycobacterium tuberculosis: testing a putative role in menaquinone biosynthesis.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3E3Q
 
 | |
3EBG
 
 | | Structure of the M1 Alanylaminopeptidase from malaria | | Descriptor: | GLYCEROL, M1 family aminopeptidase, MAGNESIUM ION, ... | | Authors: | McGowan, S, Porter, C.J, Buckle, A.M, Whisstock, J.C. | | Deposit date: | 2008-08-27 | | Release date: | 2009-01-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for the inhibition of the essential Plasmodium falciparum M1 neutral aminopeptidase
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3E5R
 
 | |
3EF6
 
 | | Crystal structure of Toluene 2,3-Dioxygenase Reductase | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, GLYCEROL, SULFATE ION, ... | | Authors: | Friemann, R, Lee, K, Brown, E.N, Gibson, D.T, Eklund, H, Ramaswamy, S. | | Deposit date: | 2008-09-08 | | Release date: | 2009-03-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the multicomponent Rieske non-heme iron toluene 2,3-dioxygenase enzyme system
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3EFG
 
 | | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913 | | Descriptor: | 1,2-ETHANEDIOL, Protein slyX homolog | | Authors: | Cuff, M.E, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-09-08 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of SlyX protein from Xanthomonas campestris pv. campestris str. ATCC 33913
TO BE PUBLISHED
|
|
3E73
 
 | | Crystal Structure of Human LanCL1 complexed with GSH | | Descriptor: | GLUTATHIONE, LanC-like protein 1, ZINC ION | | Authors: | Zhang, W, Zhu, G, Li, X, Rao, Z, Zhang, C. | | Deposit date: | 2008-08-17 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione
Genes Dev., 23, 2009
|
|
3E8D
 
 | | Crystal structures of the kinase domain of AKT2 in complex with ATP-competitive inhibitors | | Descriptor: | 4-[2-(4-amino-2,5-dihydro-1,2,5-oxadiazol-3-yl)-6-{[(1S)-3-amino-1-phenylpropyl]oxy}-1-ethyl-1H-imidazo[4,5-c]pyridin-4-yl]-2-methylbut-3-yn-2-ol, Glycogen synthase kinase-3 beta peptide, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Elkins, P.A, Smallwood, A, Ward, P. | | Deposit date: | 2008-08-19 | | Release date: | 2008-10-14 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Aminofurazans as potent inhibitors of AKT kinase
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3EHR
 
 | | Crystal Structure of Human Osteoclast Stimulating Factor | | Descriptor: | Osteoclast-stimulating factor 1 | | Authors: | Tong, S, Zhou, H, Gao, Y, Zhu, Z, Zhang, X, Teng, M, Niu, L. | | Deposit date: | 2008-09-14 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of human osteoclast stimulating factor
Proteins, 75, 2009
|
|
3EIK
 
 | |
3E94
 
 | |
3EJQ
 
 | | Golgi alpha-Mannosidase II in complex with 5-substitued swainsonine analog: (5R)-5-[2'-oxo-2'-(4-methylphenyl)ethyl]-swainsonine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, 1-(4-methylphenyl)-2-[(1S,2R,5R,8R,8aR)-1,2,8-trihydroxyoctahydroindolizin-5-yl]ethanone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A, Rose, D.R. | | Deposit date: | 2008-09-18 | | Release date: | 2009-10-13 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Investigation of the Binding of 5-Substituted Swainsonine Analogues to Golgi alpha-Mannosidase II.
Chembiochem, 11, 2010
|
|
3EA9
 
 | |
3EKW
 
 | | Crystal structure of the inhibitor Atazanavir (ATV) in complex with a multi-drug resistance HIV-1 protease variant (L10I/G48V/I54V/V64I/V82A) Refer: FLAP+ in citation. | | Descriptor: | (3S,8S,9S,12S)-3,12-BIS(1,1-DIMETHYLETHYL)-8-HYDROXY-4,11-DIOXO-9-(PHENYLMETHYL)-6-[[4-(2-PYRIDINYL)PHENYL]METHYL]-2,5, 6,10,13-PENTAAZATETRADECANEDIOIC ACID DIMETHYL ESTER, PHOSPHATE ION, ... | | Authors: | Prabu-Jeyabalan, M, King, N.M, Bandaranayake, R.M. | | Deposit date: | 2008-09-19 | | Release date: | 2009-09-01 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Extreme Entropy-Enthalpy Compensation in a Drug-Resistant Variant of HIV-1 Protease.
Acs Chem.Biol., 7, 2012
|
|
3EAY
 
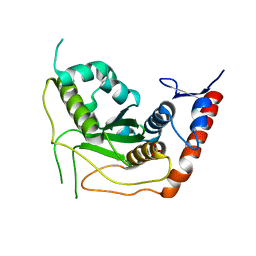 | | Crystal structure of the human SENP7 catalytic domain | | Descriptor: | SULFATE ION, Sentrin-specific protease 7 | | Authors: | Lima, C.D, Reverter, D. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Human SENP7 Catalytic Domain and Poly-SUMO Deconjugation Activities for SENP6 and SENP7.
J.Biol.Chem., 283, 2008
|
|
3EB5
 
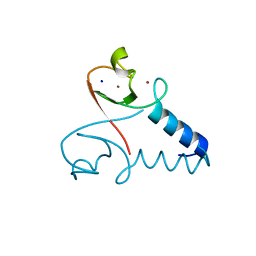 | | Structure of the cIAP2 RING domain | | Descriptor: | Baculoviral IAP repeat-containing protein 3, SODIUM ION, ZINC ION | | Authors: | Mace, P.D, Linke, K, Smith, C.A, Day, C.L. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the cIAP2 RING domain reveal conformational changes associated with ubiquitin-conjugating enzyme (E2) recruitment.
J.Biol.Chem., 283, 2008
|
|
3DJH
 
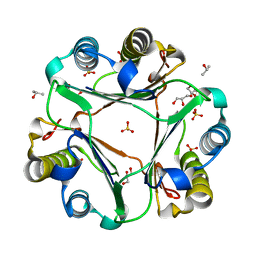 | |
3DOJ
 
 | | Structure of Glyoxylate reductase 1 from Arabidopsis (AtGLYR1) | | Descriptor: | CHLORIDE ION, Dehydrogenase-like protein | | Authors: | Jorgensen, R. | | Deposit date: | 2008-07-04 | | Release date: | 2009-07-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.101 Å) | | Cite: | Cytosolic NADPH-dependent Glyoxylate Reductase from Arabidopsis: Crystal Structure and Kinetic Characterization of Active Site Mutants
To be Published
|
|
3DOW
 
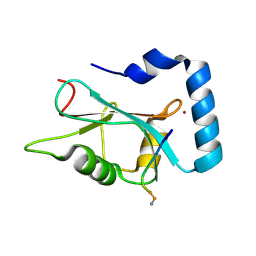 | |
3DP3
 
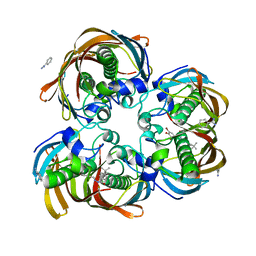 | | Crystal structure of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase (FabZ) from Helicobacter pylori in complex with compound 3q | | Descriptor: | (3R)-hydroxymyristoyl-acyl carrier protein dehydratase, 4-tert-butyl-N'-[(1E)-(3,5-dibromo-2,4-dihydroxyphenyl)methylidene]benzohydrazide, BENZAMIDINE, ... | | Authors: | Zhang, L, He, L, Liu, X, Liu, H, Shen, X, Jiang, H. | | Deposit date: | 2008-07-07 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Discovering potent inhibitors against the beta-hydroxyacyl-acyl carrier protein dehydratase (FabZ) of Helicobacter pylori: structure-based design, synthesis, bioassay, and crystal structure determination.
J.Med.Chem., 52, 2009
|
|
3DPQ
 
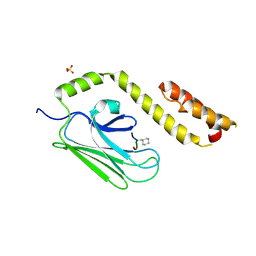 | |
3DQG
 
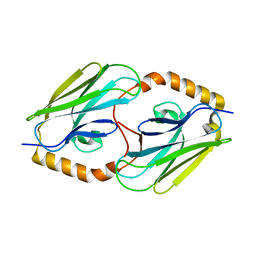 | | Peptide-binding domain of heat shock 70 kDa protein F, mitochondrial precursor, from Caenorhabditis elegans. | | Descriptor: | Heat shock 70 kDa protein F | | Authors: | Osipiuk, J, Mulligan, R, Gu, M, Voisine, C, Morimoto, R.I, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-07-09 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | X-ray crystal structure of peptide-binding domain of heat shock 70 kDa protein F, mitochondrial precursor, from Caenorhabditis elegans.
To be Published
|
|
