3QGD
 
 | | Crystal structure of the hepatitis C virus NS5B RNA-dependent RNA polymerase complex with (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid and (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-1-[(4-ethylphenyl)sulfonyl]-N-(4-methoxybenzyl)piperazine-2-carboxamide | | Descriptor: | (2E)-3-(4-{[(1-{[(13-cyclohexyl-6-oxo-6,7-dihydro-5H-indolo[1,2-d][1,4]benzodiazepin-10-yl)carbonyl]amino}cyclopentyl)carbonyl]amino}phenyl)prop-2-enoic acid, (2R)-4-(2,6-dimethoxypyrimidin-4-yl)-1-[(4-ethylphenyl)sulfonyl]-N-(4-methoxybenzyl)piperazine-2-carboxamide, RNA-directed RNA polymerase, ... | | Authors: | Sheriff, S. | | Deposit date: | 2011-01-24 | | Release date: | 2011-04-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Investigation of the mode of binding of a novel series of N-benzyl-4-heteroaryl-1-(phenylsulfonyl)piperazine-2-carboxamides to the hepatitis C virus polymerase.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4Q96
 
 | | CID of human RPRD1B in complex with an unmodified CTD peptide | | Descriptor: | RPB1-CTD, Regulation of nuclear pre-mRNA domain-containing protein 1B, SULFATE ION, ... | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-04-29 | | Release date: | 2014-06-04 | | Last modified: | 2014-08-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
3QGH
 
 | |
3QGI
 
 | |
4FLA
 
 | | Crystal structure of human RPRD1B, carboxy-terminal domain | | Descriptor: | Regulation of nuclear pre-mRNA domain-containing protein 1B, UNKNOWN ATOM OR ION | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Guo, X, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2012-06-14 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
1YSV
 
 | |
3CSO
 
 | | HCV Polymerase in complex with a 1,5 Benzodiazepine inhibitor | | Descriptor: | (11S)-10-acetyl-11-[4-(benzyloxy)-3-chlorophenyl]-3,3-dimethyl-2,3,4,5,10,11-hexahydro-1H-dibenzo[b,e][1,4]diazepin-1-one, RNA-directed RNA polymerase | | Authors: | Nyanguile, O. | | Deposit date: | 2008-04-10 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | 1,5-benzodiazepines, a novel class of hepatitis C virus polymerase nonnucleoside inhibitors.
Antimicrob.Agents Chemother., 52, 2008
|
|
3NYB
 
 | |
4Q94
 
 | | human RPRD1B CID in complex with a RPB1-CTD derived Ser2 phosphorylated peptide | | Descriptor: | Regulation of nuclear pre-mRNA domain-containing protein 1B, SULFATE ION, UNKNOWN ATOM OR ION, ... | | Authors: | Ni, Z, Xu, C, Tempel, W, El Bakkouri, M, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Greenblatt, J.F, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-04-29 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | RPRD1A and RPRD1B are human RNA polymerase II C-terminal domain scaffolds for Ser5 dephosphorylation.
Nat.Struct.Mol.Biol., 21, 2014
|
|
8R1J
 
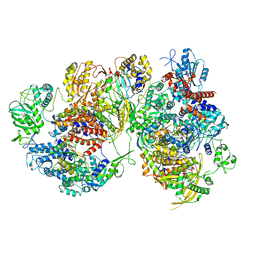 | | Structure of avian H5N1 influenza A polymerase dimer in complex with human ANP32B. | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member B, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Carrique, L, Staller, E, Keown, J.R, Fan, H, Fodor, E, Grimes, J.M. | | Deposit date: | 2023-11-02 | | Release date: | 2024-05-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structures of H5N1 influenza polymerase with ANP32B reveal mechanisms of genome replication and host adaptation.
Nat Commun, 15, 2024
|
|
2R7W
 
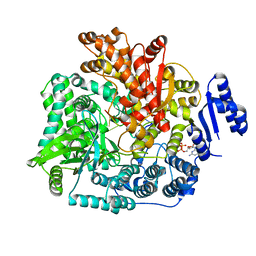 | | Crystal Structure of Rotavirus SA11 VP1/RNA (UGUGACC)/mRNA 5'-CAP (m7GpppG) complex | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, RNA (5'-R(*UP*GP*UP*GP*AP*CP*C)-3'), RNA-dependent RNA polymerase | | Authors: | Lu, X, Harrison, S.C, Tao, Y.J, Patton, J.T, Nibert, M.L. | | Deposit date: | 2007-09-10 | | Release date: | 2008-07-29 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Mechanism for coordinated RNA packaging and genome replication by rotavirus polymerase VP1.
Structure, 16, 2008
|
|
6FO1
 
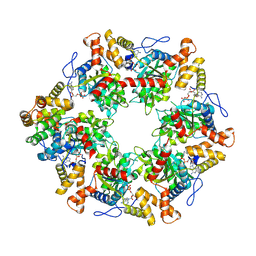 | | Human R2TP subcomplex containing 1 RUVBL1-RUVBL2 hexamer bound to 1 RBD domain from RPAP3. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RNA polymerase II-associated protein 3, RuvB-like 1, ... | | Authors: | Martino, F, Munoz-Hernandez, H, Rodriguez, C.F, Pearl, L.H, Llorca, O. | | Deposit date: | 2018-02-05 | | Release date: | 2018-04-04 | | Last modified: | 2019-12-11 | | Method: | ELECTRON MICROSCOPY (3.57 Å) | | Cite: | RPAP3 provides a flexible scaffold for coupling HSP90 to the human R2TP co-chaperone complex.
Nat Commun, 9, 2018
|
|
2XHV
 
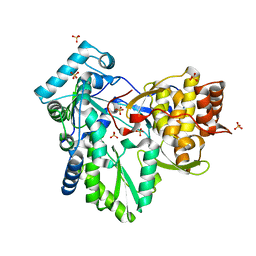 | | HCV-J4 NS5B Polymerase Point Mutant Orthorhombic Crystal Form | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase, SULFATE ION | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-21 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
2X1B
 
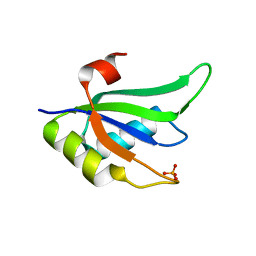 | | Structure of RNA15 RRM | | Descriptor: | MRNA 3'-END-PROCESSING PROTEIN RNA15, PHOSPHATE ION | | Authors: | Pancevac, C, Goldstone, D.C, Ramos, A, Taylor, I.A. | | Deposit date: | 2010-01-06 | | Release date: | 2010-02-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the RNA15 Rrm-RNA Complex Reveals the Molecular Basis of Gu Specificity in Transcriptional 3-End Processing Factors.
Nucleic Acids Res., 38, 2010
|
|
2XI3
 
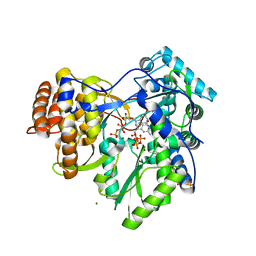 | | HCV-H77 NS5B Polymerase Complexed With GTP | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, RNA-directed RNA polymerase | | Authors: | Harrus, D, Ahmed-El-Sayed, N, Simister, P.C, Miller, S, Triconnet, M, Hagedorn, C.H, Mahias, K, Rey, F.A, Astier-Gin, T, Bressanelli, S. | | Deposit date: | 2010-06-25 | | Release date: | 2010-08-04 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Further Insights Into the Roles of GTP and the C- Terminus of the Hepatitis C Virus Polymerase in the Initiation of RNA Synthesis
J.Biol.Chem., 285, 2010
|
|
7N68
 
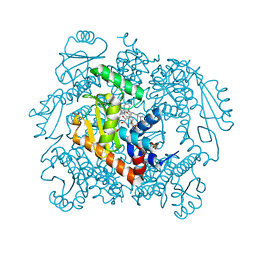 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000988288 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein,Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-06-07 | | Release date: | 2022-06-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6V6X
 
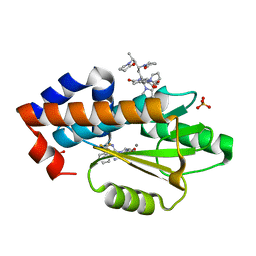 | | The crystal structure of the 2009 H1N1 PA endonuclease mutant I38T in complex with SJ000988632 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, N-[2-(6-amino-9H-purin-9-yl)ethyl]-5-hydroxy-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-06 | | Release date: | 2020-12-09 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VG9
 
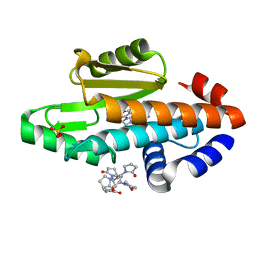 | | The crystal structure of the 2009 H1N1/California PA endonuclease I38T mutant in complex with SJ000986248 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2020-01-07 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6V9E
 
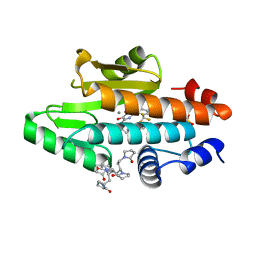 | | The crystal structure of the 2009 H1N1 PA endonuclease wild type in complex with SJ000988632 | | Descriptor: | 1,1',1'',1''',1''''-[(3R,5S,7S,9R)-decane-1,3,5,7,9-pentayl]penta(pyrrolidin-2-one), MANGANESE (II) ION, N-[2-(6-amino-9H-purin-9-yl)ethyl]-5-hydroxy-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-13 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
6VBR
 
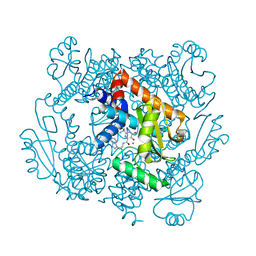 | | The crystal structure of the 2009 H1N1/California PA endonuclease wild type in complex with SJ000986248 | | Descriptor: | Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Rankovic, Z, White, S.W. | | Deposit date: | 2019-12-19 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
7MPF
 
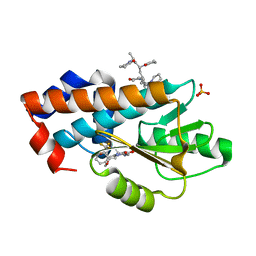 | | The crystal structure of wild type PA endonuclease (2009/H1N1/CALIFORNIA) in complex with SJ000986436 | | Descriptor: | 5-hydroxy-N-[2-(4-hydroxy-3-methoxyphenyl)ethyl]-6-oxo-2-[2-(trifluoromethyl)phenyl]-1,6-dihydropyrimidine-4-carboxamide, Hexa Vinylpyrrolidone K15, MANGANESE (II) ION, ... | | Authors: | Cuypers, M.G, Slavish, J.P, Rankovic, Z, White, S.W. | | Deposit date: | 2021-05-04 | | Release date: | 2022-05-04 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
3HKY
 
 | | HCV NS5B polymerase genotype 1b in complex with 1,5 benzodiazepine 6 | | Descriptor: | (11S)-10-[(2,5-dimethyl-1,3-oxazol-4-yl)carbonyl]-11-{2-fluoro-4-[(2-methylprop-2-en-1-yl)oxy]phenyl}-3,3-dimethyl-2,3,4,5,10,11-hexahydrothiopyrano[3,2-b][1,5]benzodiazepin-6-ol 1,1-dioxide, CHLORIDE ION, RNA-directed RNA polymerase, ... | | Authors: | Nyanguile, O, De Bondt, H.L. | | Deposit date: | 2009-05-26 | | Release date: | 2010-05-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 1a/1b subtype profiling of nonnucleoside polymerase inhibitors of hepatitis C virus
J.Virol., 84, 2010
|
|
3FQL
 
 | | Hepatitis C virus polymerase NS5B (CON1 1-570) with HCV-796 inhibitor | | Descriptor: | 5-cyclopropyl-2-(4-fluorophenyl)-6-[(2-hydroxyethyl)(methylsulfonyl)amino]-N-methyl-1-benzofuran-3-carboxamide, GLYCEROL, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2009-01-07 | | Release date: | 2009-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Slow binding inhibition and mechanism of resistance of non-nucleoside polymerase inhibitors of hepatitis C virus.
J.Biol.Chem., 284, 2009
|
|
3FQK
 
 | | Hepatitis C virus polymerase NS5B (BK 1-570) with HCV-796 inhibitor | | Descriptor: | 5-cyclopropyl-2-(4-fluorophenyl)-6-[(2-hydroxyethyl)(methylsulfonyl)amino]-N-methyl-1-benzofuran-3-carboxamide, RNA-directed RNA polymerase | | Authors: | Harris, S.F, Wong, A. | | Deposit date: | 2009-01-07 | | Release date: | 2009-02-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Slow binding inhibition and mechanism of resistance of non-nucleoside polymerase inhibitors of hepatitis C virus.
J.Biol.Chem., 284, 2009
|
|
8PTJ
 
 | |
