4DPE
 
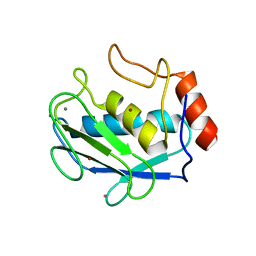 | | Structure of MMP3 complexed with a platinum-based inhibitor. | | Descriptor: | CALCIUM ION, CHLORIDE ION, N-ISOBUTYL-N-[4-METHOXYPHENYLSULFONYL]GLYCYL HYDROXAMIC ACID, ... | | Authors: | Belviso, B.D, Arnesano, F, Calderone, V, Caliandro, R, Natile, G, Siliqi, D. | | Deposit date: | 2012-02-13 | | Release date: | 2013-02-20 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure of matrix metalloproteinase-3 with a platinum-based inhibitor.
Chem.Commun.(Camb.), 49, 2013
|
|
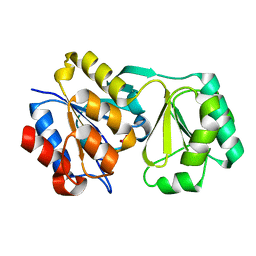
2RAR
 
 | |
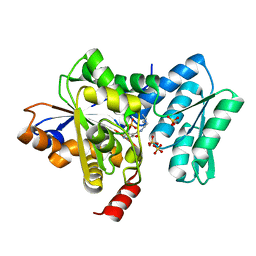
4H27
 
 | |
3FIB
 
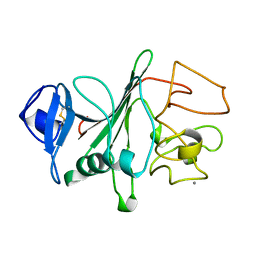 | | RECOMBINANT HUMAN GAMMA-FIBRINOGEN CARBOXYL TERMINAL FRAGMENT (RESIDUES 143-411) BOUND TO CALCIUM AT PH 6.0: A FURTHER REFINEMENT OF PDB ENTRY 1FIB, AND DIFFERS FROM 1FIB BY THE MODELLING OF A CIS PEPTIDE BOND BETWEEN RESIDUES K338 AND C339 | | Descriptor: | CALCIUM ION, FIBRINOGEN GAMMA CHAIN RESIDUES | | Authors: | Pratt, K.P, Cote, H.C.F, Chung, D.W, Stenkamp, R.E, Davie, E.W. | | Deposit date: | 1997-07-14 | | Release date: | 1997-09-17 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The primary fibrin polymerization pocket: three-dimensional structure of a 30-kDa C-terminal gamma chain fragment complexed with the peptide Gly-Pro-Arg-Pro.
Proc.Natl.Acad.Sci.USA, 94, 1997
|
|
2I9S
 
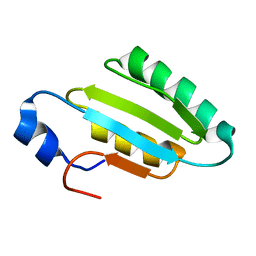 | | The solution structure of the core of mesoderm development (MESD). | | Descriptor: | Mesoderm development candidate 2 | | Authors: | Koehler, C, Andersen, O, Diehl, A, Schmieder, P, Krause, G, Oschkinat, H. | | Deposit date: | 2006-09-06 | | Release date: | 2007-05-01 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the core of mesoderm development (MESD), a chaperone for members of the LDLR-family
J.STRUCT.FUNCT.GENOM., 7, 2006
|
|
3GC7
 
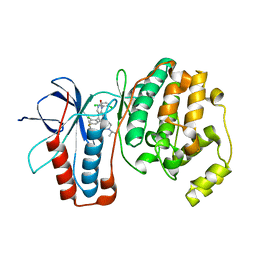 | | The structure of p38alpha in complex with a dihydroquinazolinone | | Descriptor: | 5-(2-chloro-4-fluorophenyl)-1-(2,6-dichlorophenyl)-7-[1-(1-methylethyl)piperidin-4-yl]-3,4-dihydroquinazolin-2(1H)-one, Mitogen-activated protein kinase 14 | | Authors: | Scapin, G, Patel, S.B. | | Deposit date: | 2009-02-21 | | Release date: | 2009-07-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The three-dimensional structure of MAP kinase p38beta: different features of the ATP-binding site in p38beta compared with p38alpha.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2HK7
 
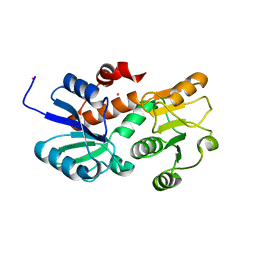 | | Crystal structure of shikimate dehydrogenase from aquifex aeolicus in complex with mercury at 2.5 angstrom resolution | | Descriptor: | MERCURY (II) ION, Shikimate dehydrogenase | | Authors: | Gan, J.H, Prabakaran, P, Gu, Y.J, Andrykovitch, M, Li, Y, Liu, H.H, Yan, H, Ji, X. | | Deposit date: | 2006-07-03 | | Release date: | 2007-06-19 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and biochemical analyses of shikimate dehydrogenase AroE from Aquifex aeolicus: implications for the catalytic mechanism.
Biochemistry, 46, 2007
|
|
3F5U
 
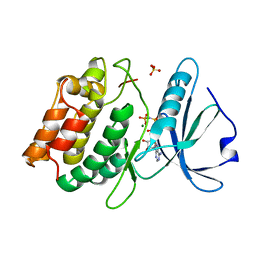 | | Crystal structure of the death associated protein kinase in complex with AMPPNP and Mg2+ | | Descriptor: | Death-associated protein kinase 1, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | McNamara, L.K, Watterson, D.M, Brunzelle, J.S. | | Deposit date: | 2008-11-04 | | Release date: | 2009-03-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural insight into nucleotide recognition by human death-associated protein kinase.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3F9Q
 
 | |
3PH9
 
 | | Crystal structure of the human anterior gradient protein 3 | | Descriptor: | Anterior gradient protein 3 homolog | | Authors: | Nguyen, V.D, Ruddock, L.W, Salin, M, Wierenga, R.K. | | Deposit date: | 2010-11-03 | | Release date: | 2011-10-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Crystal structure of human anterior gradient protein 3.
Acta Crystallogr.,Sect.F, 74, 2018
|
|
3GC9
 
 | | The structure of p38beta C119S, C162S in complex with a dihydroquinazolinone inhibitor | | Descriptor: | 5-(2-chloro-4-fluorophenyl)-1-(2,6-dichlorophenyl)-7-[1-(1-methylethyl)piperidin-4-yl]-3,4-dihydroquinazolin-2(1H)-one, Mitogen-activated protein kinase 11, SODIUM ION, ... | | Authors: | Scapin, G, Patel, S.B. | | Deposit date: | 2009-02-21 | | Release date: | 2009-07-21 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The three-dimensional structure of MAP kinase p38beta: different features of the ATP-binding site in p38beta compared with p38alpha.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
4E1B
 
 | | Re-refinement of PDB entry 2EQA - SUA5 protein from Sulfolobus tokodaii with bound threonylcarbamoyladenylate | | Descriptor: | MAGNESIUM ION, YrdC/Sua5 family protein, threonylcarbamoyladenylate | | Authors: | Parthier, C, Goerlich, S, Jaenecke, F, Breithaupt, C, Braeuer, U, Fandrich, U, Clausnitzer, D, Wehmeier, U.F, Boettcher, C, Scheel, D, Stubbs, M.T. | | Deposit date: | 2012-03-06 | | Release date: | 2012-03-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The O-Carbamoyltransferase TobZ Catalyzes an Ancient Enzymatic Reaction.
Angew.Chem.Int.Ed.Engl., 51, 2012
|
|
3HPF
 
 | | Crystal structure of the mutant Y90F of divergent galactarate dehydratase from Oceanobacillus iheyensis complexed with Mg and galactarate | | Descriptor: | D-galactaric acid, MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Rakus, J.F, Gerlt, J.A, Almo, S.C. | | Deposit date: | 2009-06-04 | | Release date: | 2009-12-22 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
1R4S
 
 | | URATE OXIDASE FROM ASPERGILLUS FLAVUS COMPLEXED WITH ITS INHIBITOR 9-METHYL URIC ACID | | Descriptor: | 9-METHYL URIC ACID, CYSTEINE, Uricase | | Authors: | Retailleau, P, Colloc'h, N, Prange, T. | | Deposit date: | 2003-10-08 | | Release date: | 2004-03-02 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Complexed and ligand-free high-resolution structures of urate oxidase (Uox) from Aspergillus flavus: a reassignment of the active-site binding mode.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
3PMO
 
 | | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, UDP-3-O-[3-hydroxymyristoyl] glucosamine N-acyltransferase | | Authors: | Badger, J, Chie-Leon, B, Logan, C, Sridhar, V, Sankaran, B, Zwart, P.H, Nienaber, V. | | Deposit date: | 2010-11-17 | | Release date: | 2011-07-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | The structure of LpxD from Pseudomonas aeruginosa at 1.3 A resolution.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
4IDW
 
 | | Polycrystalline T6 Bovine Insulin: Anisotropic Lattice Evolution and Novel Structure Refinement Strategy | | Descriptor: | Insulin A chain, Insulin B chain, ZINC ION | | Authors: | Margiolaki, I, Giannopoulou, A.E, Wright, J.P, Knight, L, Norrman, M, Schluckebier, G, Fitch, A, Von Dreele, R.B. | | Deposit date: | 2012-12-13 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-30 | | Method: | POWDER DIFFRACTION | | Cite: | High-resolution powder X-ray data reveal the T6 hexameric form of bovine insulin
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2YN8
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, STAUROSPORINE | | Authors: | Read, J, Brassington, C.A, Overmann, R. | | Deposit date: | 2012-10-13 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Stability and Solubility Engineering of the Ephb4 Tyrosine Kinase Catalytic Domain Using a Rationally Designed Synthetic Library.
Protein Eng.Des.Sel., 26, 2013
|
|
3SB2
 
 | | Crystal Structure of the RNA chaperone Hfq from Herbaspirillum seropedicae SMR1 | | Descriptor: | GLYCEROL, Protein hfq | | Authors: | Kadowaki, M.A.S, Iulek, J, Barbosa, J.A.R.G, Pedrosa, F.O, Souza, E.M, Chubatsu, L.S, Monteiro, R.A, Steffens, M.B.R. | | Deposit date: | 2011-06-03 | | Release date: | 2012-01-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6301 Å) | | Cite: | Structural characterization of the RNA chaperone Hfq from the nitrogen-fixing bacterium Herbaspirillum seropedicae SmR1.
Biochim.Biophys.Acta, 1824, 2011
|
|
3H3F
 
 | | Rabbit muscle L-lactate dehydrogenase in complex with NADH and oxamate | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, ACETATE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Bujacz, A, Bujacz, G, Swiderek, K, Paneth, P. | | Deposit date: | 2009-04-16 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Modeling of isotope effects on binding oxamate to lactic dehydrogenase
J.Phys.Chem.B, 113, 2009
|
|
4I8N
 
 | | CRYSTAL STRUCTURE of PROTEIN TYROSINE PHOSPHATASE 1B IN COMPLEX WITH AN INHIBITOR [(4-{(2S)-2-(1,3-BENZOXAZOL-2-YL)-2-[(4-FLUOROPHENYL)SULFAMOYL]ETHYL}PHENYL)AMINO](OXO)ACETIC ACID | | Descriptor: | Tyrosine-protein phosphatase non-receptor type 1, [(4-{(2S)-2-(1,3-benzoxazol-2-yl)-2-[(4-fluorophenyl)sulfamoyl]ethyl}phenyl)amino](oxo)acetic acid | | Authors: | Reddy, S.M.V.V.V, Rao, K.N, Subramanya, H. | | Deposit date: | 2012-12-03 | | Release date: | 2012-12-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | X-Ray Structure of PTP1B in Complex with a New PTP1B Inhibitor.
Protein Pept.Lett., 21, 2014
|
|
2YEX
 
 | | Synthesis and evaluation of triazolones as checkpoint kinase 1 inhibitors | | Descriptor: | 5-METHYL-8-(1H-PYRROL-2-YL)[1,2,4]TRIAZOLO[4,3-A]QUINOLIN-1(2H)-ONE, GLYCEROL, SERINE/THREONINE-PROTEIN KINASE CHK1, ... | | Authors: | Read, J.A, Breed, J, Haye, H, McCall, E, Vallentine, A, White, A. | | Deposit date: | 2011-03-31 | | Release date: | 2012-03-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Synthesis and Evaluation of Triazolones as Checkpoint Kinase 1 Inhibitors.
Bioorg.Med.Chem.Lett., 22, 2012
|
|
2GZS
 
 | |
5OFT
 
 | |
2ZT9
 
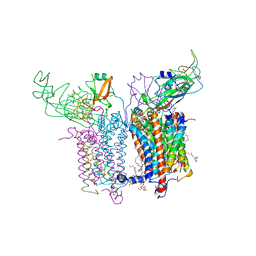 | | Crystal Structure of the Cytochrome b6f Complex from Nostoc sp. PCC 7120 | | Descriptor: | (7R,17E)-4-HYDROXY-N,N,N,7-TETRAMETHYL-7-[(8E)-OCTADEC-8-ENOYLOXY]-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOS-17-EN-1-AMINIUM 4-OXIDE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, Apocytochrome f, ... | | Authors: | Craner, W.A, Baniulis, D, Yamashita, E. | | Deposit date: | 2008-09-27 | | Release date: | 2009-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-Function, Stability, and Chemical Modification of the Cyanobacterial Cytochrome b6f Complex from Nostoc sp. PCC 7120
J.Biol.Chem., 284, 2009
|
|
3FYY
 
 | | Crystal structure of divergent enolase from Oceanobacillus iheyensis complexed with Mg | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-01-23 | | Release date: | 2009-02-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Computation-facilitated assignment of the function in the enolase superfamily: a regiochemically distinct galactarate dehydratase from Oceanobacillus iheyensis .
Biochemistry, 48, 2009
|
|
