6TVB
 
 | | Crystal structure of the haemagglutinin from a transmissible H10N7 seal influenza virus isolated in Netherland in complex with human receptor analogue 6'-SLN | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Hemagglutinin HA1, ... | | Authors: | Zhang, J, Xiong, X, Purkiss, A, Walker, P, Gamblin, S, Skehel, J.J. | | Deposit date: | 2020-01-09 | | Release date: | 2020-10-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Hemagglutinin Traits Determine Transmission of Avian A/H10N7 Influenza Virus between Mammals.
Cell Host Microbe, 28, 2020
|
|
9ART
 
 | | Crystal structure of SARS-CoV-2 main protease A191T mutant in complex with an inhibitor 5h | | Descriptor: | 3C-like proteinase nsp5, N-[(2S)-1-({(1S,2S)-1-(1,3-benzothiazol-2-yl)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}amino)-4-methyl-1-oxopentan-2-yl]-4-methoxy-1H-indole-2-carboxamide | | Authors: | Bulut, H, Hattori, S, Hayashi, H, Hasegawa, K, Li, M, Wlodawer, A, Tamamura, H, Mitsuya, H. | | Deposit date: | 2024-02-23 | | Release date: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structural and virologic mechanism of emergence of main protease inhibitor-resistance in SARS-CoV-2 as selected with main protease inhibitors
To Be Published
|
|
8WIX
 
 | | cryo-EM structure of alligator haemoglobin in carbonmonoxy form | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Takahashi, K, Lee, Y, Fago, A, Bautista, N.M, Kawamoto, A, Kurisu, G, Storz, J, Nishizawa, T, Tame, J.R.H. | | Deposit date: | 2023-09-25 | | Release date: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.29 Å) | | Cite: | The unique allosteric property of crocodilian haemoglobin elucidated by cryo-EM.
Nat Commun, 15, 2024
|
|
8XE7
 
 | |
3UO4
 
 | | Aurora A in complex with RPM1680 | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-(biphenyl-2-ylamino)pyrimidin-2-yl]amino}benzoic acid, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-11-16 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | A Novel Mechanism by Which Small Molecule Inhibitors Induce the DFG Flip in Aurora A.
Acs Chem.Biol., 7, 2012
|
|
8VDW
 
 | |
8YQ4
 
 | | Structure of mBaoJin2 | | Descriptor: | CHLORIDE ION, GLYCEROL, SULFATE ION, ... | | Authors: | Boyko, K.M, Nikolaeva, A.Y, Minyaev, M.E, Kuzmicheva, T.P, Vlaskina, A.V, Popov, V.O, Pyatkevich, K.D, Subach, F.V. | | Deposit date: | 2024-03-19 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of mBaoJin2
To Be Published
|
|
8Y1J
 
 | |
9LDT
 
 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
8VEN
 
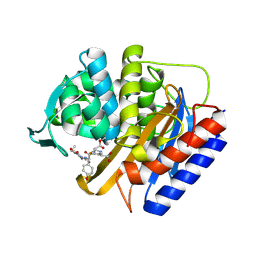 | | Crystal structure of transpeptidase domain of PBP2 from Neisseria gonorrhoeae cephalosporin-resistant strain H041 in complex with cefoperazone | | Descriptor: | (2R,4R)-2-[(1R)-1-{[(2R)-2-[(4-ethyl-2,3-dioxopiperazine-1-carbonyl)amino]-2-(4-hydroxyphenyl)acetyl]amino}-2-oxoethyl]-5-methylidene-1,3-thiazinane-4-carboxylic acid, Probable peptidoglycan D,D-transpeptidase PenA | | Authors: | Stratton, C, Bala, S, Davies, C. | | Deposit date: | 2023-12-20 | | Release date: | 2024-03-20 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ureidopenicillins Are Potent Inhibitors of Penicillin-Binding Protein 2 from Multidrug-Resistant Neisseria gonorrhoeae H041.
Acs Infect Dis., 10, 2024
|
|
9BRW
 
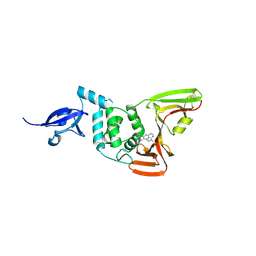 | | SARS-CoV-2 Papain-like Protease (PLpro) with Fragment 7 | | Descriptor: | CHLORIDE ION, N-(2-chlorophenyl)-1-methyl-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Papain-like protease nsp3, ... | | Authors: | Amporndanai, K, Zhao, B, Fesik, S.W. | | Deposit date: | 2024-05-11 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Fragment-Based Screen of SARS-CoV-2 Papain-like Protease (PLpro)
Acs Med.Chem.Lett., 2024
|
|
8WOV
 
 | | Crystal structure of Arabidopsis thaliana UDP-glucose 4-epimerase 2 (AtUGE2) complexed with UDP, G233A mutant | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase 2, URIDINE-5'-DIPHOSPHATE | | Authors: | Matsumoto, M, Umezawa, A, Kotake, T, Fushinobu, S. | | Deposit date: | 2023-10-07 | | Release date: | 2024-05-15 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Cytosolic UDP-L-arabinose synthesis by bifunctional UDP-glucose 4-epimerases in Arabidopsis.
Plant J., 119, 2024
|
|
8Z10
 
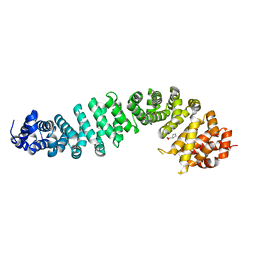 | | Human beta-catenin crystal structure | | Descriptor: | Catenin beta-1, PROLINE | | Authors: | Tim, F. | | Deposit date: | 2024-04-10 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of Beta-catenin
To Be Published
|
|
9FT0
 
 | | Yeast 20S proteasome in complex with epoxyketone inhibitor 42 | | Descriptor: | (2S)-N-[(2S)-1-[[(1S)-2-cyclohexyl-1-[(2R,3S,6R,7S)-3-methanoyl-2,6-dimethyl-6,7-bis(oxidanyl)-1,4-oxazepan-7-yl]ethyl]amino]-3-(4-methoxyphenyl)-1-oxidanylidene-propan-2-yl]-2-(2-morpholin-4-ylethanoylamino)-4-oxidanyl-butanamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, ... | | Authors: | Maurits, E, Huber, E.M, Dekker, P.M, Wang, X, Heinemeyer, W, Florea, B.I, Groll, M, Overkleeft, H.S. | | Deposit date: | 2024-06-23 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-based design of peptide epoxyketones selectively targeting the three human immunoproteasome active sites
to be published
|
|
9BKS
 
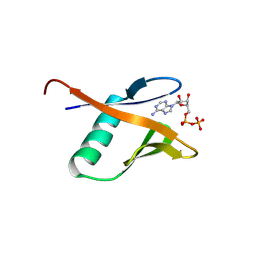 | | Crystal structure of the Human TRIP12 WWE domain (isoform 2) in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Isoform 2 of E3 ubiquitin-protein ligase TRIP12 | | Authors: | Kimani, S, Dong, A, Li, Y, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-04-29 | | Release date: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | Crystal structure of the Human TRIP12 WWE domain (isoform 2) in complex with ADP
To be published
|
|
8WYP
 
 | |
9LDB
 
 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
8WY3
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor 21 | | Descriptor: | Bromodomain-containing protein 4, ~{N}-cyclopropyl-2-[[5-[2-(4-fluoranyl-2,6-dimethyl-phenoxy)-5-(2-oxidanylpropan-2-yl)phenyl]-1-methyl-2-oxidanylidene-pyridin-4-yl]amino]ethanamide | | Authors: | Xu, H, Zhao, X, Shen, H, Xu, Y, Wu, X. | | Deposit date: | 2023-10-30 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Discovery of Novel Phenoxyaryl Pyridones as Bromodomain and Extra-Terminal Domain (BET) Inhibitors with High Selectivity for the Second Bromodomain (BD2) to Potentially Treat Acute Myeloid Leukemia.
J.Med.Chem., 67, 2024
|
|
9FA3
 
 | | Gcase in complex with small molecule inhibitor 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Lysosomal acid glucosylceramidase, ... | | Authors: | Tisi, D, Cleasby, A. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Fragment-Based Discovery of a Series of Allosteric-Binding Site Modulators of beta-Glucocerebrosidase.
J.Med.Chem., 67, 2024
|
|
8W3V
 
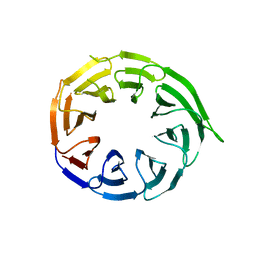 | | Crystal structure of human WDR41 | | Descriptor: | WD repeat-containing protein 41 | | Authors: | Hutchinson, A, Dong, A, Li, Y, Seitova, A, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Halabelian, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2024-02-22 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of human WDR41
To be published
|
|
3UP7
 
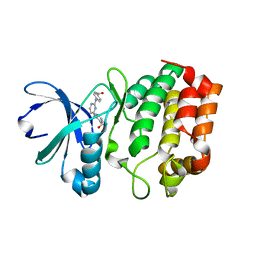 | | Aurora A in complex with YL1-038-09 | | Descriptor: | 2-({2-[(4-carboxyphenyl)amino]pyrimidin-4-yl}amino)benzoic acid, Aurora kinase A | | Authors: | Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2011-11-17 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Development of o-Chlorophenyl Substituted Pyrimidines as Exceptionally Potent Aurora Kinase Inhibitors.
J.Med.Chem., 55, 2012
|
|
8VL7
 
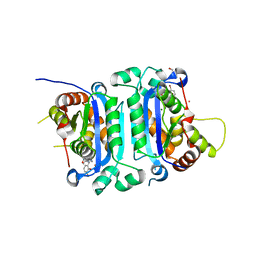 | | Co-crystal structure of human TREX1 in complex with an inhibitor | | Descriptor: | (2P)-2-[3-bromo-2-(2-hydroxyethoxy)phenyl]-5-hydroxy-1-methyl-N-(1,2-oxazol-4-yl)-6-oxo-1,6-dihydropyrimidine-4-carboxamide, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Dehghani-Tafti, S, Dong, A, Li, Y, Ackloo, S, Arrowsmith, C.H, Edwards, A.M, Halabelian, L. | | Deposit date: | 2024-01-11 | | Release date: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Co-crystal structure of human TREX1 in complex with an inhibitor
To be published
|
|
9FOF
 
 | |
9FAL
 
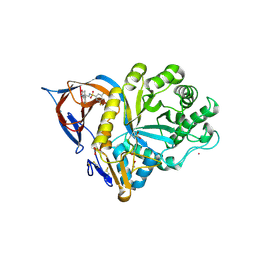 | | Gcase in complex with small molecule inhibitor 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Tisi, D, Cleasby, A. | | Deposit date: | 2024-05-10 | | Release date: | 2024-07-03 | | Last modified: | 2024-07-24 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Fragment-Based Discovery of a Series of Allosteric-Binding Site Modulators of beta-Glucocerebrosidase.
J.Med.Chem., 67, 2024
|
|
9ATX
 
 | |
