8JDM
 
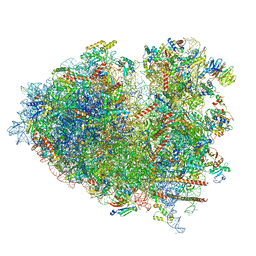 | | Structure of the Human cytoplasmic Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAU) (rotated state) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2023-05-14 | | Release date: | 2023-12-06 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.67 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
7QGG
 
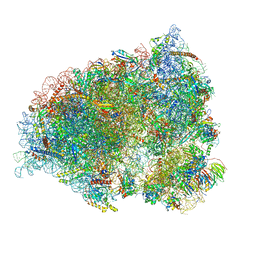 | | Neuronal RNA granules are ribosome complexes stalled at the pre-translocation state | | Descriptor: | 40S ribosomal protein S10, 40S ribosomal protein S11, 40S ribosomal protein S12, ... | | Authors: | Pulk, A, Kipper, K, Mansour, A. | | Deposit date: | 2021-12-08 | | Release date: | 2022-10-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.86 Å) | | Cite: | Neuronal RNA granules are ribosome complexes stalled at the pre-translocation state.
J.Mol.Biol., 434, 2022
|
|
8JDK
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Asp(ManQ34) and mRNA(GAU) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2023-05-14 | | Release date: | 2023-12-06 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.26 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
8JDL
 
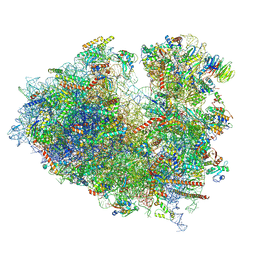 | | Structure of the Human cytoplasmic Ribosome with human tRNA Tyr(GalQ34) and mRNA(UAU) (non-rotated state) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2023-05-14 | | Release date: | 2023-12-06 | | Last modified: | 2025-02-12 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
9C3H
 
 | | Structure of the CNOT3-bound human 80S ribosome with tRNA-ARG in the P-site. | | Descriptor: | 18S rRNA, 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 28S rRNA, ... | | Authors: | Erzberger, J.P, Cruz, V.E. | | Deposit date: | 2024-06-01 | | Release date: | 2024-12-04 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | Specific tRNAs promote mRNA decay by recruiting the CCR4-NOT complex to translating ribosomes.
Science, 386, 2024
|
|
8P4F
 
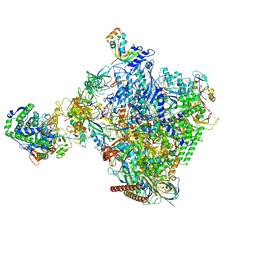 | | Structural insights into human co-transcriptional capping - structure 6 | | Descriptor: | 7N-METHYL-8-HYDROGUANOSINE-5'-TRIPHOSPHATE, Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (38-MER), ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8P4E
 
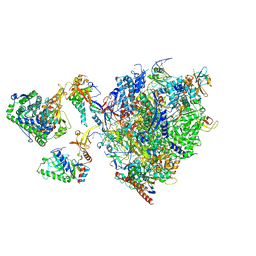 | | Structural insights into human co-transcriptional capping - structure 5 | | Descriptor: | Cap-specific mRNA (nucleoside-2'-O-)-methyltransferase 1, DNA (26-MER), DNA (35-MER), ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-07-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
6Z6M
 
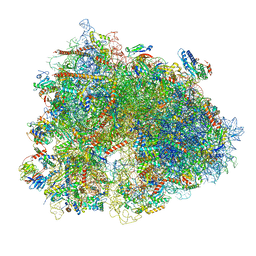 | | Cryo-EM structure of human 80S ribosomes bound to EBP1, eEF2 and SERBP1 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
6Z6N
 
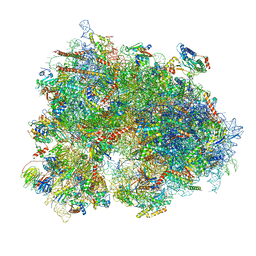 | | Cryo-EM structure of human EBP1-80S ribosomes (focus on EBP1) | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wells, J.N, Buschauer, R, Mackens-Kiani, T, Best, K, Kratzat, H, Berninghausen, O, Becker, T, Cheng, J, Beckmann, R. | | Deposit date: | 2020-05-28 | | Release date: | 2020-07-29 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure and function of yeast Lso2 and human CCDC124 bound to hibernating ribosomes.
Plos Biol., 18, 2020
|
|
8P4B
 
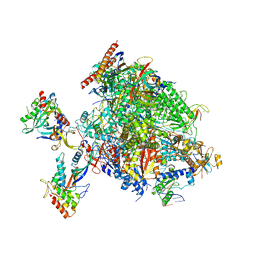 | | Structural insights into human co-transcriptional capping - structure 2 | | Descriptor: | DNA (35-MER), DNA (5'-D(P*AP*GP*CP*GP*GP*CP*GP*GP*AP*GP*CP*CP*AP*GP*CP*AP*GP*GP*GP*AP*GP*CP*TP*G)-3'), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8P4C
 
 | | Structural insights into human co-transcriptional capping - structure 3 | | Descriptor: | DNA (32-MER), DNA (41-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8P4A
 
 | | Structural insights into human co-transcriptional capping - structure 1 | | Descriptor: | DNA (29-MER), DNA (38-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
8P4D
 
 | | Structural insights into human co-transcriptional capping - structure 4 | | Descriptor: | DNA (32-MER), DNA (41-MER), DNA-directed RNA polymerase II subunit E, ... | | Authors: | Garg, G, Dienemann, C, Farnung, L, Schwarz, J, Linden, A, Urlaub, H, Cramer, P. | | Deposit date: | 2023-05-20 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into human co-transcriptional capping.
Mol.Cell, 83, 2023
|
|
7ABG
 
 | | Human pre-Bact-1 spliceosome | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, 7-METHYL-GUANOSINE-5'-TRIPHOSPHATE-5'-GUANOSINE, Cell division cycle 5-like protein, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-07 | | Release date: | 2020-12-23 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
9GUX
 
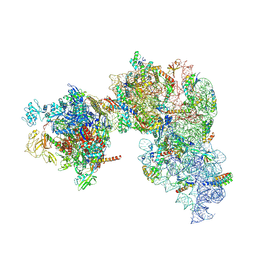 | | 30S-TEC (TEC in expressome position) Inactive state 1 | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Rahil, H, Weixlbaumer, A, Webster, M.W. | | Deposit date: | 2024-09-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Molecular basis of mRNA delivery to the bacterial ribosome.
Science, 386, 2024
|
|
9GUW
 
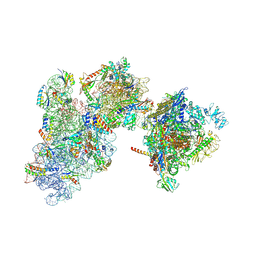 | | 30S-TEC (TEC in expressome position) Inactive state 2 | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S1, 30S ribosomal protein S10, ... | | Authors: | Rahil, H, Weixlbaumer, A, Webster, M.W. | | Deposit date: | 2024-09-20 | | Release date: | 2024-11-20 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Molecular basis of mRNA delivery to the bacterial ribosome.
Science, 386, 2024
|
|
8QOI
 
 | | Structure of the human 80S ribosome at 1.9 A resolution - the molecular role of chemical modifications and ions in RNA | | Descriptor: | 18S rRNA (1740-MER), 28S rRNA (3773-MER), 40S ribosomal protein S10, ... | | Authors: | Holvec, S, Barchet, C, Frechin, L, Hazemann, I, von Loeffelholz, O, Klaholz, B.P. | | Deposit date: | 2023-09-29 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-28 | | Method: | ELECTRON MICROSCOPY (1.9 Å) | | Cite: | The structure of the human 80S ribosome at 1.9 angstrom resolution reveals the molecular role of chemical modifications and ions in RNA.
Nat.Struct.Mol.Biol., 31, 2024
|
|
5MS0
 
 | |
6XZQ
 
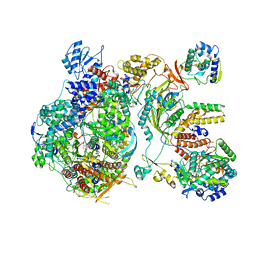 | | Influenza C virus polymerase in complex with human ANP32A - Subclass 1 | | Descriptor: | Acidic leucine-rich nuclear phosphoprotein 32 family member A, Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Fan, H, Carrique, L, Keown, J.R, Grimes, J.M, Fodor, E. | | Deposit date: | 2020-02-05 | | Release date: | 2020-11-25 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Host ANP32A mediates the assembly of the influenza virus replicase.
Nature, 587, 2020
|
|
6X43
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - II state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewellyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-22 | | Release date: | 2021-02-03 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6X50
 
 | | Mfd-bound E.coli RNA polymerase elongation complex - V state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DNA (64-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Llewelyn, E, Chen, J, Kang, J.Y, Darst, S.A. | | Deposit date: | 2020-05-24 | | Release date: | 2021-02-03 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis for transcription complex disruption by the Mfd translocase.
Elife, 10, 2021
|
|
6XH8
 
 | | CueR-transcription activation complex with RNA transcript | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
6XH7
 
 | | CueR-TAC without RNA | | Descriptor: | COPPER (II) ION, DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Liu, B, Shi, W, Yang, Y. | | Deposit date: | 2020-06-18 | | Release date: | 2021-04-14 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural basis of copper-efflux-regulator-dependent transcription activation.
Iscience, 24, 2021
|
|
3S24
 
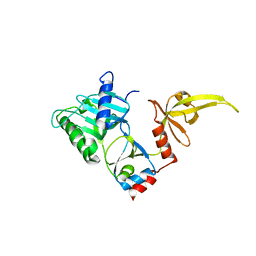 | | Crystal structure of human mRNA guanylyltransferase | | Descriptor: | SULFATE ION, mRNA-capping enzyme | | Authors: | Das, K, Chu, C, Thyminski, J.R, Bauman, J.D, Guan, R, Qiu, W, Montelione, G.T, Arnold, E, Shatkin, A.J. | | Deposit date: | 2011-05-16 | | Release date: | 2011-06-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.0137 Å) | | Cite: | Structure of the guanylyltransferase domain of human mRNA capping enzyme.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
6PB4
 
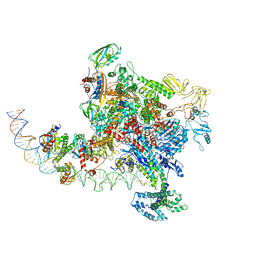 | |
