5VPX
 
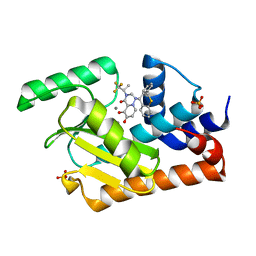 | | I38T mutant of 2009 H1N1 PA Endonuclease in complex with RO-7 | | Descriptor: | 1-[(11S)-6,11-dihydrodibenzo[b,e]thiepin-11-yl]-5-hydroxy-3-[(2R)-1,1,1-trifluoropropan-2-yl]-2,3-dihydro-1H-pyrido[2,1-f][1,2,4]triazine-4,6-dione, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-05-06 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Identification of the I38T PA Substitution as a Resistance Marker for Next-Generation Influenza Virus Endonuclease Inhibitors.
MBio, 9, 2018
|
|
5VQN
 
 | | E119D mutant of 2009 H1N1 PA Endonuclease in complex with RO-7 | | Descriptor: | 1-[(11S)-6,11-dihydrodibenzo[b,e]thiepin-11-yl]-5-hydroxy-3-[(2R)-1,1,1-trifluoropropan-2-yl]-2,3-dihydro-1H-pyrido[2,1-f][1,2,4]triazine-4,6-dione, MANGANESE (II) ION, Polymerase acidic protein, ... | | Authors: | Kumar, G, White, S.W. | | Deposit date: | 2017-05-09 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | Identification of the I38T PA Substitution as a Resistance Marker for Next-Generation Influenza Virus Endonuclease Inhibitors.
MBio, 9, 2018
|
|
6AJE
 
 | | Crystal structure of DHODH in complex with ferulenol from Eimeria tenella | | Descriptor: | 4-oxidanyl-3-[(2~{E},6~{E})-3,7,11-trimethyldodeca-2,6,10-trienyl]chromen-2-one, Dihydroorotate dehydrogenase (quinone), mitochondrial, ... | | Authors: | Shiba, T, Inaoka, D.K, Sato, D, Hartuti, E.D, Amalia, E, Nagahama, M, Yoshioka, Y, Matsubayashi, M, Balogun, E.O, Tsuji, N, Kita, K, Harada, S. | | Deposit date: | 2018-08-27 | | Release date: | 2019-08-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Structural and Biochemical Features of Eimeria tenella Dihydroorotate Dehydrogenase, a Potential Drug Target.
Genes (Basel), 11, 2020
|
|
7ZJ4
 
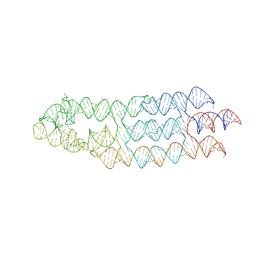 | | Ligand bound state of a brocolli-pepper aptamer FRET tile | | Descriptor: | 4-(3,5-difluoro-4-hydroxybenzyl)-1,2-dimethyl-1H-imidazol-5-ol, 4-[(~{Z})-1-cyano-2-[5-[2-hydroxyethyl(methyl)amino]thieno[3,2-b]thiophen-2-yl]ethenyl]benzenecarbonitrile, POTASSIUM ION, ... | | Authors: | McRae, E.K.S, Vallina, N.S, Hansen, B.K, Boussebayle, A, Andersen, E.S. | | Deposit date: | 2022-04-08 | | Release date: | 2023-04-19 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (4.43 Å) | | Cite: | Structure determination of Pepper-Broccoli FRET pair by RNA origami scaffolding
To Be Published
|
|
6VNT
 
 | | Tryptophan synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and sodium ion at the metal coordination site at 1.25 Angstrom resolution | | Descriptor: | 1,2-ETHANEDIOL, 2-({[4-(TRIFLUOROMETHOXY)PHENYL]SULFONYL}AMINO)ETHYL DIHYDROGEN PHOSPHATE, 2-{[(E)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}prop-2-enoic acid, ... | | Authors: | Hilario, E, Fan, L, Dunn, M.F, Mueller, L.J. | | Deposit date: | 2020-01-29 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Tryptophan synthase in complex with inhibitor N-(4'-trifluoromethoxybenzenesulfonyl)-2-amino-1-ethylphosphate (F9F) at the alpha-site, aminoacrylate at the beta site, and sodium ion at the metal coordination site at 1.25 Angstrom resolution.
To be Published
|
|
5JE5
 
 | | Crystal structure of Burkholderia glumae ToxA with bound S-adenosylhomocysteine (SAH) and 1-demethyltoxoflavin | | Descriptor: | 6-methylpyrimido[5,4-e][1,2,4]triazine-5,7(6H,8H)-dione, Methyl transferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.564 Å) | | Cite: | Burkholderia glumae ToxA Is a Dual-Specificity Methyltransferase That Catalyzes the Last Two Steps of Toxoflavin Biosynthesis.
Biochemistry, 55, 2016
|
|
5FKA
 
 | | Crystal structure of staphylococcal enterotoxin E in complex with a T cell receptor | | Descriptor: | STAPHYLOCOCCAL ENTEROTOXIN E, T CELL RECEPTOR ALPHA CHAIN, T CELL RECEPTOR BETA CHAIN, ... | | Authors: | Rodstrom, K.E.J, Regenthal, P, Lindkvist-Petersson, K. | | Deposit date: | 2015-10-15 | | Release date: | 2016-05-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Two Common Structural Motifs for Tcr Recognition by Staphylococcal Enterotoxins.
Sci.Rep., 6, 2016
|
|
8JZE
 
 | | PSI-AcpPCI supercomplex from Symbiodinium | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z.H, Li, X.Y, Wang, W.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-28 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.99 Å) | | Cite: | Structures and organizations of PSI-AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JW0
 
 | | PSI-AcpPCI supercomplex from Amphidinium carterae | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, Z.H, Li, X.Y, Wang, W.D. | | Deposit date: | 2023-06-28 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures and organizations of PSI-AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
8JZF
 
 | | PSI-AcpPCI supercomplex from Symbiodinium | | Descriptor: | (3S,3'R,5R,6S,7cis)-7',8'-didehydro-5,6-dihydro-5,6-epoxy-beta,beta-carotene-3,3'-diol, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, ... | | Authors: | Li, X.Y, Li, Z.H, Wang, W.D. | | Deposit date: | 2023-07-05 | | Release date: | 2024-02-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Structures and organizations of PSI-AcpPCI supercomplexes from red tidal and coral symbiotic photosynthetic dinoflagellates.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
7U2J
 
 | | Crystal structure of the Thermus thermophilus 70S ribosome in complex with mRNA, aminoacylated A-site Gly-NH-tRNAgly, peptidyl P-site fMAC-NH-tRNAmet, deacylated E-site tRNAgly, and chloramphenicol at 2.55A resolution | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Syroegin, E.A, Aleksandrova, E.V, Polikanov, Y.S. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-13 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for the inability of chloramphenicol to inhibit peptide bond formation in the presence of A-site glycine.
Nucleic Acids Res., 50, 2022
|
|
8OWI
 
 | |
5JE0
 
 | | Crystal structure of Burkholderia glumae ToxA with bound S-adenosylhomocysteine (SAH) and 1,6-didemethyltoxoflavin | | Descriptor: | Methyl transferase, S-ADENOSYL-L-HOMOCYSTEINE, pyrimido[5,4-e][1,2,4]triazine-5,7(6H,8H)-dione | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.552 Å) | | Cite: | Burkholderia glumae ToxA Is a Dual-Specificity Methyltransferase That Catalyzes the Last Two Steps of Toxoflavin Biosynthesis.
Biochemistry, 55, 2016
|
|
9B0T
 
 | | Cryo-EM structure of E227Q variant of uMtCK1 in complex with transition state analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase U-type, mitochondrial, ... | | Authors: | Demir, M, Koepping, L, Zhao, J, Sergienko, E. | | Deposit date: | 2024-03-12 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Structural basis for substrate binding, catalysis, and inhibition of cancer target mitochondrial creatine kinase by a covalent inhibitor.
Structure, 33, 2025
|
|
9B14
 
 | | Cryo-EM structure of human uMtCK1 in complex with transition state analog | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase U-type, mitochondrial, ... | | Authors: | Demir, M, Koepping, L, Zhao, J, Sergienko, E. | | Deposit date: | 2024-03-13 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Structural basis for substrate binding, catalysis, and inhibition of cancer target mitochondrial creatine kinase by a covalent inhibitor.
Structure, 33, 2025
|
|
9B0U
 
 | | Cryo-EM structure of E227Q variant of uMtCK1 incubated with ADP and phosphocreatine at pH 8.0 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Creatine kinase U-type, mitochondrial, ... | | Authors: | Demir, M, Koepping, L, Zhao, J, Sergienko, E. | | Deposit date: | 2024-03-12 | | Release date: | 2025-02-12 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (2.44 Å) | | Cite: | Structural basis for substrate binding, catalysis, and inhibition of cancer target mitochondrial creatine kinase by a covalent inhibitor.
Structure, 33, 2025
|
|
5OHG
 
 | | enolase in complex with RNase E | | Descriptor: | Enolase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Du, D, Luisi, B.F. | | Deposit date: | 2017-07-16 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Analysis of the natively unstructured RNA/protein-recognition core in the Escherichia coli RNA degradosome and its interactions with regulatory RNA/Hfq complexes.
Nucleic Acids Res., 46, 2018
|
|
5JE1
 
 | | Crystal structure of Burkholderia glumae ToxA with bound S-adenosylhomocysteine (SAH) and toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, Methyl transferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Burkholderia glumae ToxA Is a Dual-Specificity Methyltransferase That Catalyzes the Last Two Steps of Toxoflavin Biosynthesis.
Biochemistry, 55, 2016
|
|
5JDY
 
 | | Crystal structure of Burkholderia glumae ToxA Y7F mutant with bound S-adenosylhomocysteine (SAH) and toxoflavin | | Descriptor: | 1,6-dimethylpyrimido[5,4-e][1,2,4]triazine-5,7(1H,6H)-dione, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Methyl transferase, ... | | Authors: | Fenwick, M.K, Philmus, B, Begley, T.P, Ealick, S.E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-05-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Burkholderia glumae ToxA Is a Dual-Specificity Methyltransferase That Catalyzes the Last Two Steps of Toxoflavin Biosynthesis.
Biochemistry, 55, 2016
|
|
6U7B
 
 | | Structure of E. coli MS115-1 CdnC:HORMA-deltaN complex | | Descriptor: | ACETATE ION, ADENOSINE MONOPHOSPHATE, CHLORIDE ION, ... | | Authors: | Ye, Q, Corbett, K.D. | | Deposit date: | 2019-09-02 | | Release date: | 2019-12-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | HORMA Domain Proteins and a Trip13-like ATPase Regulate Bacterial cGAS-like Enzymes to Mediate Bacteriophage Immunity.
Mol.Cell, 77, 2020
|
|
4PUO
 
 | | Crystal structure of HIV-1 reverse transcriptase in complex with RNA/DNA and Nevirapine | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, 5'-D(*A*CP*AP*GP*TP*CP*CP*CP*TP*GP*TP*TP*CP*GP*GP*GP*CP*GP*CP*CP*G)-3', 5'-R(P*AP*UP*GP*GP*UP*CP*GP*GP*CP*GP*CP*CP*CP*GP*AP*AP*CP*AP*GP*GP*GP*AP*CP*UP*GP*UP*G)-3', ... | | Authors: | Das, K, Martinez, S.E, Arnold, E. | | Deposit date: | 2014-03-13 | | Release date: | 2014-06-18 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structures of HIV-1 RT-RNA/DNA ternary complexes with dATP and nevirapine reveal conformational flexibility of RNA/DNA: insights into requirements for RNase H cleavage.
Nucleic Acids Res., 42, 2014
|
|
4LRG
 
 | | Structure of BRD4 bromodomain 1 with a dimethyl thiophene isoxazole azepine carboxamide | | Descriptor: | 2-[(6S)-4-(4-chlorophenyl)-2,3,9-trimethyl-6H-[1,2]oxazolo[5,4-c]thieno[2,3-e]azepin-6-yl]acetamide, Bromodomain-containing protein 4 | | Authors: | Ravichandran, S, Jayaram, H, Poy, F, Gehling, V, Hewitt, M, Vaswani, R, Leblanc, Y, Cote, A, Nasveschuk, C, Taylor, A, Harmange, J.-C, Audia, J, Pardo, E, Joshi, S, Sandy, P, Mertz, J, Sims, R, Bergeron, L, Bryant, B, Yellapuntala, S, Nandana, B.S, Birudukota, S, Albrecht, B, Bellon, S. | | Deposit date: | 2013-07-19 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Discovery, Design, and Optimization of Isoxazole Azepine BET Inhibitors.
ACS Med Chem Lett, 4, 2013
|
|
6FDQ
 
 | |
1LWF
 
 | | CRYSTAL STRUCTURE OF A MUTANT HIV-1 REVERSE TRANSCRIPTASE (RTMQ+M184V: M41L/D67N/K70R/M184V/T215Y) IN COMPLEX WITH NEVIRAPINE | | Descriptor: | 11-CYCLOPROPYL-5,11-DIHYDRO-4-METHYL-6H-DIPYRIDO[3,2-B:2',3'-E][1,4]DIAZEPIN-6-ONE, HIV-1 REVERSE TRANSCRIPTASE | | Authors: | Ren, J, Chamberlain, P.P, Nichols, C.E, Douglas, L, Stuart, D.I, Stammers, D.K. | | Deposit date: | 2002-05-31 | | Release date: | 2002-10-30 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structures of Zidovudine- or Lamivudine-resistant human immunodeficiency virus type 1 reverse transcriptases containing mutations at codons 41, 184, and 215.
J.Virol., 76, 2002
|
|
7F30
 
 | | Crystal structure of OxdB E85A in complex with Z-2- (3-bromophenyl) propanal oxime | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, Phenylacetaldoxime dehydratase, Z-2-(3-bromophenyl) propanal oxime | | Authors: | Muraki, N, Matsui, D, Asano, Y, Aono, S. | | Deposit date: | 2021-06-15 | | Release date: | 2022-04-27 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structural analysis of aldoxime dehydratase from Bacillus sp. OxB-1: Importance of surface residues in optimization for crystallization.
J.Inorg.Biochem., 230, 2022
|
|
