2MKM
 
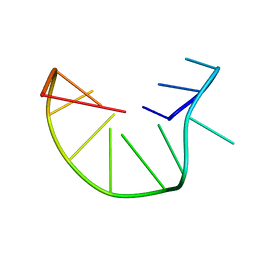 | | G-triplex structure and formation propensity | | Descriptor: | DNA_(5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*G)-3') | | Authors: | Cerofolini, L, Fragai, M, Giachetti, A, Limongelli, V, Luchinat, C, Novellino, E, Parrinello, M, Randazzo, A. | | Deposit date: | 2014-02-10 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-triplex structure and formation propensity.
Nucleic Acids Res., 42, 2014
|
|
2M4J
 
 | | 40-residue beta-amyloid fibril derived from Alzheimer's disease brain | | Descriptor: | Amyloid beta A4 protein | | Authors: | Lu, J, Qiang, W, Meredith, S.C, Yau, W, Schweiters, C.D, Tycko, R. | | Deposit date: | 2013-02-05 | | Release date: | 2013-09-25 | | Last modified: | 2024-05-15 | | Method: | SOLID-STATE NMR | | Cite: | Molecular Structure of beta-Amyloid Fibrils in Alzheimer's Disease Brain Tissue.
Cell(Cambridge,Mass.), 154, 2013
|
|
1IRL
 
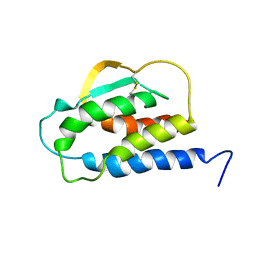 | | THE SOLUTION STRUCTURE OF THE F42A MUTANT OF HUMAN INTERLEUKIN 2 | | Descriptor: | INTERLEUKIN-2 | | Authors: | Mott, H.R, Baines, B.S, Hall, R.M, Cooke, R.M, Driscoll, P.C, Weir, M.P, Campbell, I.D. | | Deposit date: | 1995-08-25 | | Release date: | 1995-12-07 | | Last modified: | 2021-11-03 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the F42A mutant of human interleukin 2.
J.Mol.Biol., 247, 1995
|
|
1ITL
 
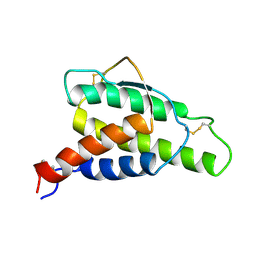 | | HUMAN INTERLEUKIN 4: THE SOLUTION STRUCTURE OF A FOUR-HELIX-BUNDLE PROTEIN | | Descriptor: | INTERLEUKIN-4 | | Authors: | Smith, L.J, Redfield, C, Boyd, J, Lawrence, G.M.P, Edwards, R.G, Smith, R.A.G, Dobson, C.M. | | Deposit date: | 1992-02-08 | | Release date: | 1993-04-15 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Human interleukin 4. The solution structure of a four-helix bundle protein.
J.Mol.Biol., 224, 1992
|
|
1IVA
 
 | |
2MKO
 
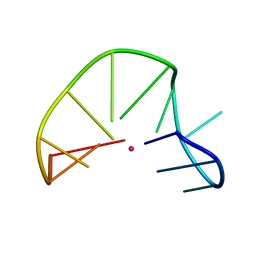 | | G-triplex structure and formation propensity | | Descriptor: | DNA_(5'-D(*GP*GP*TP*TP*GP*GP*TP*GP*TP*GP*G)-3'), POTASSIUM ION | | Authors: | Cerofolini, L, Fragai, M, Giachetti, A, Limongelli, V, Luchinat, C, Novellino, E, Parrinello, M, Randazzo, A. | | Deposit date: | 2014-02-11 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | G-triplex structure and formation propensity.
Nucleic Acids Res., 42, 2014
|
|
1JAB
 
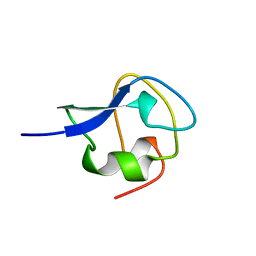 | | TYPE III ANTIFREEZE PROTEIN ISOFORM HPLC 12 T18S | | Descriptor: | PROTEIN (ANTIFREEZE PROTEIN TYPE III) | | Authors: | Graether, S.P, Deluca, C.I, Baardsnes, J, Hill, G.A, Davies, P.L, Jia, Z. | | Deposit date: | 1999-01-24 | | Release date: | 1999-04-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Quantitative and qualitative analysis of type III antifreeze protein structure and function.
J.Biol.Chem., 274, 1999
|
|
1JAF
 
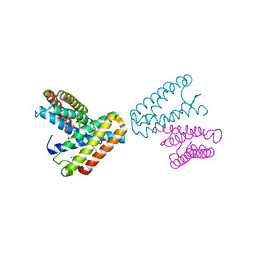 | | CRYSTAL STRUCTURE OF CYTOCHROME C' FROM RHODOCYCLUS GELATINOSUS AT 2.5 ANGSTOMS RESOLUTION | | Descriptor: | CYTOCHROME C', PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Archer, M, Banci, L, Dikaya, E, Romao, M.J. | | Deposit date: | 1997-06-24 | | Release date: | 1998-01-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Cytochrome C' from Rhodocyclus Gelatinosus and Comparison with Other Cytochromes C'
J.Biol.Inorg.Chem., 2, 1997
|
|
2M3M
 
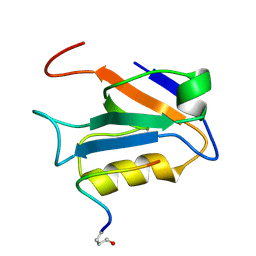 | |
1JEI
 
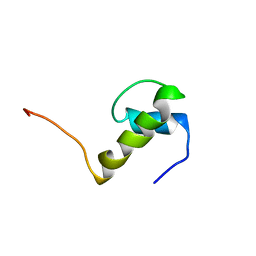 | | LEM DOMAIN OF HUMAN INNER NUCLEAR MEMBRANE PROTEIN EMERIN | | Descriptor: | EMERIN | | Authors: | Wolff, N, Gilquin, B, Courchay, K, Callebaut, I, Zinn-Justin, S. | | Deposit date: | 2001-06-18 | | Release date: | 2001-07-04 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of emerin, an inner nuclear membrane protein mutated in X-linked Emery-Dreifuss muscular dystrophy
FEBS LETT., 501, 2001
|
|
2LXE
 
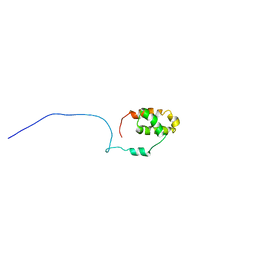 | | S4wyild | | Descriptor: | Histone-lysine N-methyltransferase SUVR4 | | Authors: | Kristiansen, P, Rahman, M.A, Aalen, R.B. | | Deposit date: | 2012-08-20 | | Release date: | 2013-11-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The arabidopsis histone methyltransferase SUVR4 binds ubiquitin via a domain with a four-helix bundle structure.
Biochemistry, 53, 2014
|
|
2M0N
 
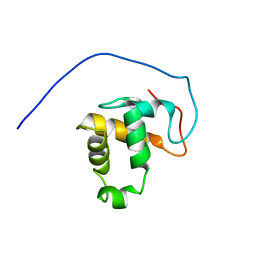 | |
2M4Z
 
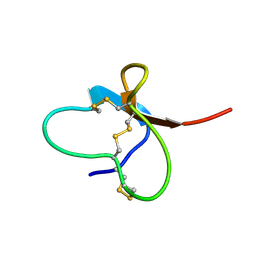 | |
2MCE
 
 | |
1K3K
 
 | | Solution Structure of a Bcl-2 Homolog from Kaposi's Sarcoma Virus | | Descriptor: | functional anti-apoptotic factor vBCL-2 homolog | | Authors: | Huang, Q, Petros, A.M, Virgin, H.W, Fesik, S.W, Olejniczak, E.T. | | Deposit date: | 2001-10-03 | | Release date: | 2002-04-10 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a Bcl-2 homolog from Kaposi sarcoma virus.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
2N0W
 
 | | Mdmx-SJ212 | | Descriptor: | 4-({(4S,5R)-4-(5-bromo-2-fluorophenyl)-5-(4-chlorophenyl)-2-[4-methoxy-2-(propan-2-yloxy)phenyl]-4,5-dihydro-1H-imidazol-1-yl}carbonyl)piperazin-2-one, Protein Mdm4 | | Authors: | Grace, C.R, Kriwacki, R.W. | | Deposit date: | 2015-03-17 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Monitoring Ligand-Induced Protein Ordering in Drug Discovery.
J.Mol.Biol., 428, 2016
|
|
2N1W
 
 | |
2N06
 
 | | Mdmx-298 | | Descriptor: | 4-[[(4S,5R)-5-(4-chlorophenyl)-4-(3-methoxyphenyl)-2-(4-methoxy-2-propan-2-yloxy-phenyl)-4,5-dihydroimidazol-1-yl]carbonyl]piperazin-2-one, Protein Mdm4 | | Authors: | Grace, C.R, Kriwacki, R.W. | | Deposit date: | 2015-03-04 | | Release date: | 2016-01-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Monitoring Ligand-Induced Protein Ordering in Drug Discovery.
J.Mol.Biol., 428, 2016
|
|
2MPV
 
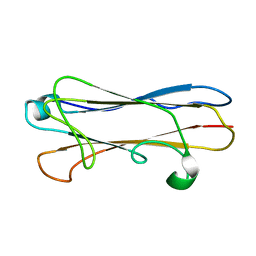 | | Structural insight into host recognition and biofilm formation by aggregative adherence fimbriae of enteroaggregative Esherichia coli | | Descriptor: | Major fimbrial subunit of aggregative adherence fimbria II AafA | | Authors: | Matthews, S.J, Yang, Y, Berry, A.A, Pakharukova, N, Garnett, J.A, Lee, W, Cota, E, Liu, B, Roy, S, Tuittila, M, Marchant, J, Inman, K.G, Ruiz-Perez, F, Mandomando, I, Nataro, J.P, Zavialov, A.V. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural insight into host recognition by aggregative adherence fimbriae of enteroaggregative Escherichia coli.
Plos Pathog., 10, 2014
|
|
2N0Q
 
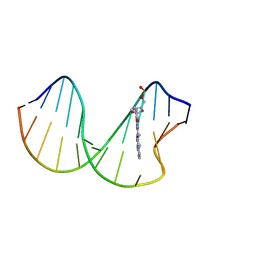 | | N2-dG-IQ modified DNA at the G1 position of the NarI recognition sequence | | Descriptor: | DNA_(5'-D(*CP*TP*CP*(IQG)P*GP*CP*GP*CP*CP*AP*TP*C)-3'), DNA_(5'-D(*GP*AP*TP*GP*GP*CP*GP*CP*CP*GP*AP*G)-3') | | Authors: | Stavros, K, Hawkins, E, Rizzo, C, Stone, M. | | Deposit date: | 2015-03-11 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Base-Displaced Intercalated Conformation of the 2-Amino-3-methylimidazo[4,5-f]quinoline N(2)-dG DNA Adduct Positioned at the Nonreiterated G(1) in the NarI Restriction Site.
Chem.Res.Toxicol., 28, 2015
|
|
1HMA
 
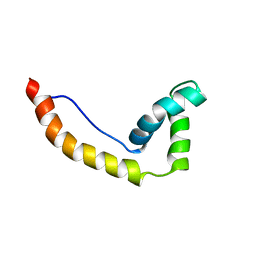 | | THE SOLUTION STRUCTURE AND DYNAMICS OF THE DNA BINDING DOMAIN OF HMG-D FROM DROSOPHILA MELANOGASTER | | Descriptor: | HMG-D | | Authors: | Jones, D.N.M, Searles, M.A, Shaw, G.L, Churchill, M.E.A, Ner, S.S, Keeler, J, Travers, A.A, Neuhaus, D. | | Deposit date: | 1994-05-12 | | Release date: | 1994-07-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure and dynamics of the DNA-binding domain of HMG-D from Drosophila melanogaster.
Structure, 2, 1994
|
|
1HZL
 
 | | SOLUTION STRUCTURES OF C-1027 APOPROTEIN AND ITS COMPLEX WITH THE AROMATIZED CHROMOPHORE | | Descriptor: | C-1027 APOPROTEIN, C-1027 AROMATIZED CHROMOPHORE | | Authors: | Tanaka, T, Fukuda-Ishisaka, S, Hirama, M, Otani, T. | | Deposit date: | 2001-01-25 | | Release date: | 2001-05-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Solution structures of C-1027 apoprotein and its complex with the aromatized chromophore.
J.Mol.Biol., 309, 2001
|
|
1KX9
 
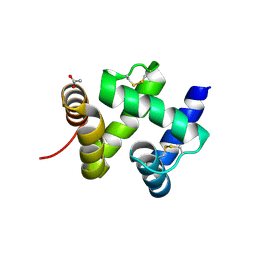 | | ANTENNAL CHEMOSENSORY PROTEIN A6 FROM THE MOTH MAMESTRA BRASSICAE | | Descriptor: | ACETATE ION, CHEMOSENSORY PROTEIN A6 | | Authors: | Lartigue, A, Campanacci, V, Roussel, A, Larsson, A.M, Jones, T.A, Tegoni, M, Cambillau, C. | | Deposit date: | 2002-01-31 | | Release date: | 2002-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | X-ray structure and ligand binding study of a moth chemosensory protein
J.Biol.Chem., 277, 2002
|
|
2MWK
 
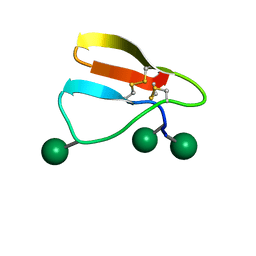 | | Family 1 Carbohydrate-Binding Module from Trichoderma reesei Cel7A with O-mannose residues at Thr1, Ser3, and Ser14 | | Descriptor: | Exoglucanase 1, alpha-D-mannopyranose | | Authors: | Happs, R.M, Chen, L, Resch, M.G, Davis, M.F, Beckham, G.T, Tan, Z, Crowley, M.F. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-02 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | O-glycosylation effects on family 1 carbohydrate-binding module solution structures.
Febs J., 282, 2015
|
|
2MWJ
 
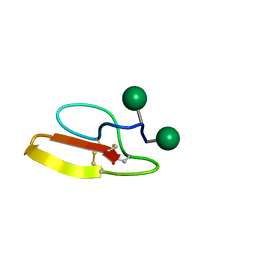 | | Solution structure of Family 1 Carbohydrate-Binding Module from Trichoderma reesei Cel7A with O-mannose residues at Thr1 and Ser3 | | Descriptor: | Exoglucanase 1, alpha-D-mannopyranose | | Authors: | Happs, R.M, Chen, L, Resch, M.G, Davis, M.F, Beckham, G.T, Tan, Z, Crowley, M.F. | | Deposit date: | 2014-11-12 | | Release date: | 2015-09-02 | | Last modified: | 2020-07-29 | | Method: | SOLUTION NMR | | Cite: | O-glycosylation effects on family 1 carbohydrate-binding module solution structures.
Febs J., 282, 2015
|
|
