5OBV
 
 | | Mycoplasma genitalium DnaK deletion mutant lacking SBDalpha in complex with ADP and Pi. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Chaperone protein DnaK, PHOSPHATE ION | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
5OBU
 
 | | Mycoplasma genitalium DnaK deletion mutant lacking SBDalpha in complex with AMPPNP. | | Descriptor: | Chaperone protein DnaK, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Adell, M, Calisto, B, Fita, I, Martinelli, L. | | Deposit date: | 2017-06-29 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The nucleotide-bound/substrate-bound conformation of the Mycoplasma genitalium DnaK chaperone.
Protein Sci., 27, 2018
|
|
5O4P
 
 | | Crystal structure of AMPylated GRP78 | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE MONOPHOSPHATE, SULFATE ION | | Authors: | Yan, Y, Chen, R, Ron, D, Read, R. | | Deposit date: | 2017-05-30 | | Release date: | 2017-11-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | AMPylation targets the rate-limiting step of BiP's ATPase cycle for its functional inactivation.
Elife, 6, 2017
|
|
5NRO
 
 | | Structure of full-length DnaK with bound J-domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Chaperone protein DnaJ, Chaperone protein DnaK, ... | | Authors: | Kopp, J, Kityk, R, Mayer, M.P. | | Deposit date: | 2017-04-25 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Molecular Mechanism of J-Domain-Triggered ATP Hydrolysis by Hsp70 Chaperones.
Mol. Cell, 69, 2018
|
|
5MKS
 
 | | HSP72-NBD bound to compound TCI 8 - Tyr15 in down-conformation | | Descriptor: | 3-[(2~{R},3~{S},4~{R},5~{R})-5-[6-azanyl-8-[(4-chlorophenyl)methylamino]purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]propyl prop-2-enoate, Heat shock 70 kDa protein 1A | | Authors: | Pettinger, J, Westwood, I.M, Cronin, N, Le Bihan, Y.-V, Van Montfort, R.L.M. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | An Irreversible Inhibitor of HSP72 that Unexpectedly Targets Lysine-56.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MKR
 
 | | HSP72-NBD bound to compound TCI 8 - Tyr15 in up-conformation | | Descriptor: | 3-[(2~{R},3~{S},4~{R},5~{R})-5-[6-azanyl-8-[(4-chlorophenyl)methylamino]purin-9-yl]-3,4-bis(oxidanyl)oxolan-2-yl]propyl prop-2-enoate, CITRATE ANION, Heat shock 70 kDa protein 1A | | Authors: | Pettinger, J, Westwood, I.M, Cronin, N, Le Bihan, Y.-V, Van Montfort, R.L.M. | | Deposit date: | 2016-12-05 | | Release date: | 2017-03-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | An Irreversible Inhibitor of HSP72 that Unexpectedly Targets Lysine-56.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5MB9
 
 | | Crystal structure of the eukaryotic ribosome associated complex (RAC), a unique Hsp70/Hsp40 pair | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Gumiero, A, Weyer, F.A, Valentin Gese, G, Lapouge, K, Sinning, I. | | Deposit date: | 2016-11-07 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into a unique Hsp70-Hsp40 interaction in the eukaryotic ribosome-associated complex.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5GJJ
 
 | |
5FPN
 
 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand 3,5-dimethyl-1H-pyrazole-4-carboxylic acid (AT9084) in an alternate binding site. | | Descriptor: | 3,5-DIMETHYL-1H-PYRAZOLE-4-CARBOXYLIC ACID, HEAT SHOCK-RELATED 70 KDA PROTEIN 2 | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FPM
 
 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand 5-phenyl-1,3,4-oxadiazole-2-thiol (AT809) in an alternate binding site. | | Descriptor: | 5-PHENYL-1,3,4-OXADIAZOLE-2-THIOL, HEAT SHOCK-RELATED 70KDA PROTEIN 2 | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-12-02 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FPE
 
 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand 1H-1,2,4-triazol-3-amine (AT485) in an alternate binding site. | | Descriptor: | 3-AMINO-1,2,4-TRIAZOLE, HEAT SHOCK-RELATED 70KDA PROTEIN 2 | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-28 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5FPD
 
 | | Structure of heat shock-related 70kDA protein 2 with small-molecule ligand pyrazine-2-carboxamide (AT513) in an alternate binding site. | | Descriptor: | HEAT SHOCK-RELATED 70KDA PROTEIN 2, PYRAZINE-2-CARBOXAMIDE | | Authors: | Jhoti, H, Ludlow, R.F, Patel, S, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-28 | | Release date: | 2015-12-16 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5F2R
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with AMP-PCP | | Descriptor: | 78 kDa glucose-regulated protein, MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-12-02 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5F1X
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with ATP | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-30 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5F0X
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 2'-deoxy-ADP and inorganic phosphate | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 2'-DEOXYADENOSINE-5'-DIPHOSPHATE, 78 kDa glucose-regulated protein, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-28 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5EY4
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 2'-deoxy-ATP | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, 78 kDa glucose-regulated protein | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-24 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5EXW
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 7-deaza-ATP | | Descriptor: | 7-deazaadenosine-5'-triphosphate, 78 kDa glucose-regulated protein | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-24 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5EX5
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with 7-deaza-ADP and inorganic phosphate | | Descriptor: | 7-deazaadenosine-5'-diphosphate, 78 kDa glucose-regulated protein, MAGNESIUM ION, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-23 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5EVZ
 
 | | Crystal structure of human GRP78 (70kDa heat shock protein 5 / BIP) ATPase domain in complex with ADP and inorganic phosphate | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Hughes, S.J, Antoshchenko, T, Song, J.H, Pizarro, J, Park, H.W. | | Deposit date: | 2015-11-20 | | Release date: | 2016-06-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Probing the ATP Site of GRP78 with Nucleotide Triphosphate Analogs.
Plos One, 11, 2016
|
|
5E86
 
 | | isolated SBD of BiP with loop34 modification | | Descriptor: | 78 kDa glucose-regulated protein | | Authors: | Liu, Q, Yang, J, Nune, M, Zong, Y, Zhou, L. | | Deposit date: | 2015-10-13 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.681 Å) | | Cite: | Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP.
Structure, 23, 2015
|
|
5E85
 
 | | isolated SBD of BiP | | Descriptor: | 78 kDa glucose-regulated protein, SULFATE ION | | Authors: | Liu, Q, Yang, J, Nune, M, Zong, Y, Zhou, L. | | Deposit date: | 2015-10-13 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP.
Structure, 23, 2015
|
|
5E84
 
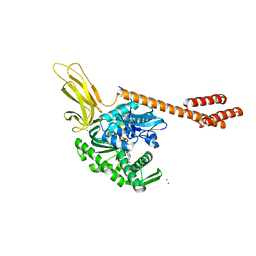 | | ATP-bound state of BiP | | Descriptor: | 78 kDa glucose-regulated protein, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Liu, Q, Yang, J, Nune, M, Zong, Y, Zhou, L. | | Deposit date: | 2015-10-13 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Close and Allosteric Opening of the Polypeptide-Binding Site in a Human Hsp70 Chaperone BiP.
Structure, 23, 2015
|
|
5BPN
 
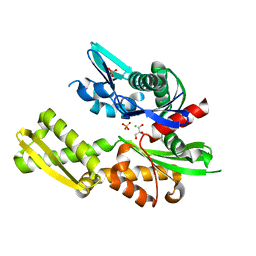 | |
5BPM
 
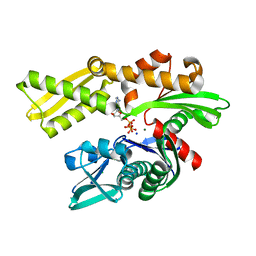 | |
5BPL
 
 | |
