8U8Q
 
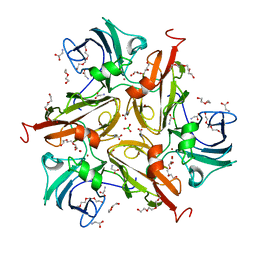 | | V290N/S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8U8P
 
 | | S292F Streptomyces coelicolor Laccase | | Descriptor: | BORIC ACID, COPPER (II) ION, Copper oxidase, ... | | Authors: | Wang, J.-X, Lu, Y. | | Deposit date: | 2023-09-18 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Increasing Reduction Potentials of Type 1 Copper Center and Catalytic Efficiency of Small Laccase from Streptomyces coelicolor through Secondary Coordination Sphere Mutations.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8U5H
 
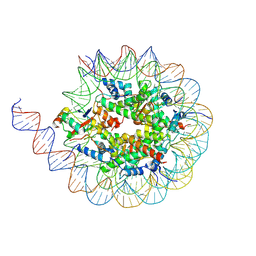 | |
8U5E
 
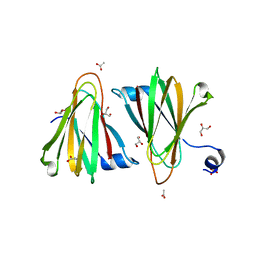 | |
8U5D
 
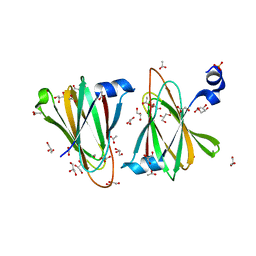 | |
8U2M
 
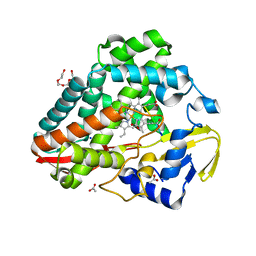 | |
8U1W
 
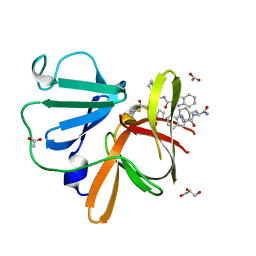 | | Structure of Norovirus (Hu/GII.4/Sydney/NSW0514/2012/AU) protease bound to inhibitor NV-004 | | Descriptor: | ACETATE ION, GLYCEROL, Peptidase C37, ... | | Authors: | Eruera, A.R, Campbell, A.C, Krause, K.L. | | Deposit date: | 2023-09-03 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Crystal Structure of Inhibitor-Bound GII.4 Sydney 2012 Norovirus 3C-Like Protease.
Viruses, 15, 2023
|
|
8U14
 
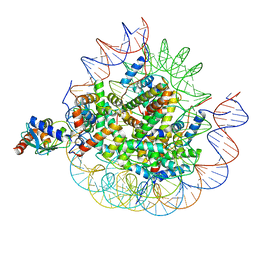 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A lysine 15 in complex with RNF168-UbcH5c (class 2) | | Descriptor: | DNA (146-MER), DNA (147-MER), E3 ubiquitin-protein ligase RNF168, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-30 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8U13
 
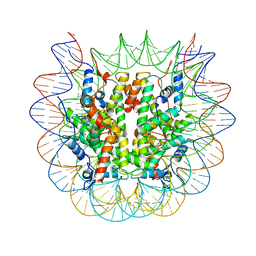 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A lysine 15 in complex with RNF168-UbcH5c (class 1) | | Descriptor: | DNA (146-MER), DNA (147-MER), E3 ubiquitin-protein ligase RNF168, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-30 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8U01
 
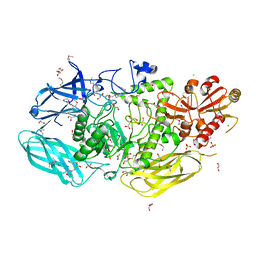 | | Crystal Structure of the Glycoside Hydrolase Family 2 TIM Barrel-domain Containing Protein from Phocaeicola plebeius | | Descriptor: | 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETATE ION, ... | | Authors: | Kim, Y, Joachimiak, G, Endres, M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2023-08-28 | | Release date: | 2023-09-27 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of the Glycoside Hydrolase Family 2 TIM Barrel-domain
Containing Protein from Phocaeicola plebeius
To Be Published
|
|
8TXX
 
 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 3) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXW
 
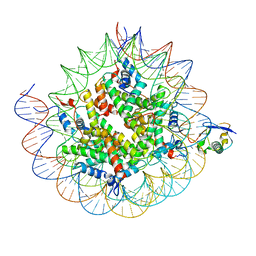 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 2) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TXV
 
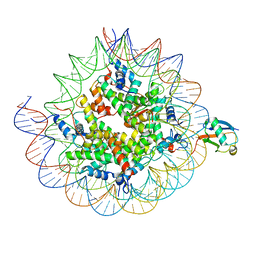 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 1) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8TWD
 
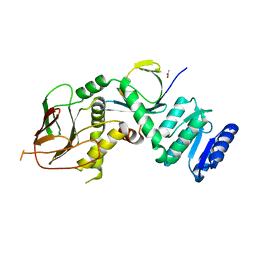 | | Structure of IraM-bound RssB | | Descriptor: | ACETATE ION, Anti-adapter protein IraM, Regulator of RpoS | | Authors: | Brugger, C, Deaconescu, A.M. | | Deposit date: | 2023-08-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | IraM remodels the RssB segmented helical linker to stabilize sigma s against degradation by ClpXP.
J.Biol.Chem., 300, 2023
|
|
8TS5
 
 | | Structure of the apo FabS1C_C1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L.L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-08-10 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8TOF
 
 | | Rpd3S bound to an H3K36Cme3 modified nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (176-MER), Histone H2A, ... | | Authors: | Markert, J.W, Vos, S.M, Farnung, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the complete Saccharomyces cerevisiae Rpd3S-nucleosome complex.
Nat Commun, 14, 2023
|
|
8TNS
 
 | |
8THU
 
 | |
8TFG
 
 | |
8TFF
 
 | | PorX with BeF3 phosphate analog | | Descriptor: | ACETATE ION, BERYLLIUM TRIFLUORIDE ION, CALCIUM ION, ... | | Authors: | Saran, A, Zeytuni, N. | | Deposit date: | 2023-07-11 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | PorX with BeF3 phosphate analog
To Be Published
|
|
8TED
 
 | |
8TCD
 
 | | Structure of Alistipes sp. 3-Keto-beta-glucopyranoside-1,2-Lyase AL1 | | Descriptor: | ACETATE ION, COBALT (II) ION, GLYCEROL, ... | | Authors: | Lazarski, A.C, Worrall, L.J, Strynadka, N.C.J. | | Deposit date: | 2023-06-30 | | Release date: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An alternative broad-specificity pathway for glycan breakdown in bacteria
Nature, 2024
|
|
8TB5
 
 | | TYK2 JH2 bound to Compound7 | | Descriptor: | ACETATE ION, N-{(3P)-3-[3-(dimethylsulfamoyl)phenyl]-1H-pyrrolo[2,3-c]pyridin-5-yl}cyclopropanecarboxamide, Non-receptor tyrosine-protein kinase TYK2 | | Authors: | Argiriadi, M.A, Van Epps, S.A, Breinlinger, E.C. | | Deposit date: | 2023-06-28 | | Release date: | 2023-10-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Targeting the Tyrosine Kinase 2 (TYK2) Pseudokinase Domain: Discovery of the Selective TYK2 Inhibitor ABBV-712.
J.Med.Chem., 66, 2023
|
|
8T9H
 
 | |
8T9F
 
 | |
