8TIW
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with blunt ends and 3' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-3', DNA (5'-D(*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Tuning chemical DNA ligation within DNA crystals and co-crystals.
To Be Published
|
|
8TIV
 
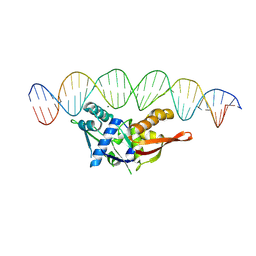 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with blunt ends and 5' terminal phosphates and crosslinked with EDC. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3')|, DNA (5'-D(A*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Tuning chemical DNA ligation within DNA crystals and co-crystals.
To Be Published
|
|
8TIU
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 2 sticky base overhangs and 3' terminal phosphates. | | Descriptor: | DNA (5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-3')|, DNA (5'-D(GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Tuning chemical DNA ligation within DNA crystals and co-crystals.
To Be Published
|
|
8TIT
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 2 sticky base overhangs and no terminal phosphates. | | Descriptor: | DNA (5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3')|, DNA (5'-D(GP*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Tuning chemical DNA ligation within DNA crystals and co-crystals.
To Be Published
|
|
8TIS
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 1 sticky bases and 3' terminal phosphates. | | Descriptor: | DNA (5'-D(GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*A)-3'), DNA(5'-D(CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-')|, MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Tuning chemical DNA ligation within DNA crystals and co-crystals.
To Be Published
|
|
8TIR
 
 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with 1 sticky bases and 5' terminal phosphates. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3'), DNA (5'-D(A*GP*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Shrestha, R, Slaughter, C.K, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Tuning chemical DNA ligation within DNA crystals and co-crystals.
To Be Published
|
|
8TIQ
 
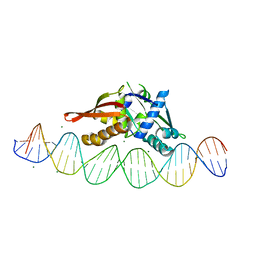 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with blunt ends and 3' terminal phosphates. | | Descriptor: | DNA (5'-D(*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*GP*A)-3', DNA (5'-D(*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*GP*A)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Slaughter, C.K, Shrestha, R, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Tuning chemical DNA ligation within DNA crystals and protein-DNA co-crystals
To Be Published
|
|
8TIP
 
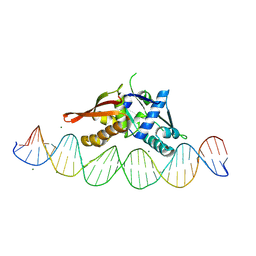 | | Isoreticular, interpenetrating co-crystal of Replication Initiator Protein REPE54 and symmetrical expanded duplex (31mer) containing the cognate REPE54 sequence and an additional G-C rich sequence with blunt ends and 5' terminal phosphates. | | Descriptor: | DNA (5'-D(A*CP*CP*CP*GP*GP*AP*CP*CP*TP*GP*TP*GP*AP*CP*AP*AP*AP*TP*TP*GP*CP*CP*CP*TP*CP*AP*GP*AP*CP*GP*G)-3')|, DNA (5'-D(A*CP*CP*GP*TP*CP*TP*GP*AP*GP*GP*GP*CP*AP*AP*TP*TP*TP*GP*TP*CP*AP*CP*AP*GP*GP*TP*CP*CP*GP*GP*G)-3'), MAGNESIUM ION, ... | | Authors: | Orun, A.R, Shields, E.T, Slaughter, C.K, Shrestha, R, Snow, C.D. | | Deposit date: | 2023-07-20 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Tuning chemical DNA ligation within DNA crystals and co-crystals.
To Be Published
|
|
8TIC
 
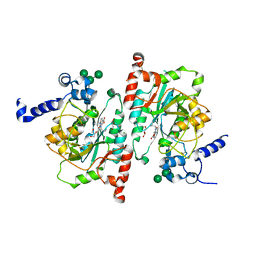 | | Structure of human beta 1,3-N-acetylglucosaminyltransferase 2 with compound 1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(2-methylpropyl)-2-oxo-2,3-dihydro-1H-benzimidazole-5-carboxylic acid, CHLORIDE ION, ... | | Authors: | Sudom, A, Min, X. | | Deposit date: | 2023-07-19 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Imidazolone as an Amide Bioisostere in the Development of beta-1,3- N -Acetylglucosaminyltransferase 2 (B3GNT2) Inhibitors.
J.Med.Chem., 66, 2023
|
|
8TI4
 
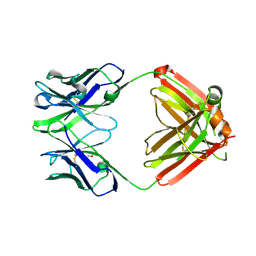 | | monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the CH1-CL interface | | Descriptor: | GLYCEROL, IgG1 Fab heavy chain, mutated to promote correct pairing, ... | | Authors: | Oganesyan, V.Y, van Dyk, N, Mazor, Y. | | Deposit date: | 2023-07-19 | | Release date: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Robust production of monovalent bispecific IgG antibodies through novel electrostatic steering mutations at the C H 1-C lambda interface.
Mabs, 15, 2023
|
|
8TI2
 
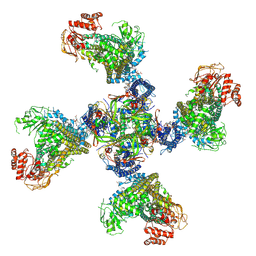 | |
8TI1
 
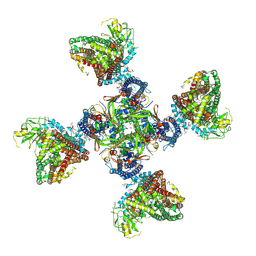 | |
8THV
 
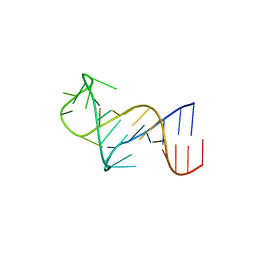 | | FARFAR-NMR ensemble of HIV-1 TAR with apical loop capturing ground and excited conformational states | | Descriptor: | RNA (29-MER) | | Authors: | Roy, R, Geng, A, Shi, H, Merriman, D.K, Dethoff, E.A, Salmon, L, Al-Hashimi, H.M. | | Deposit date: | 2023-07-18 | | Release date: | 2023-08-02 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Kinetic Resolution of the Atomic 3D Structures Formed by Ground and Excited Conformational States in an RNA Dynamic Ensemble.
J.Am.Chem.Soc., 145, 2023
|
|
8THP
 
 | |
8THN
 
 | | KcsA M96V mutant with Y78ester in High K+ | | Descriptor: | KcsA Fab Heavy Chain, KcsA Fab Light Chain, POTASSIUM ION, ... | | Authors: | Reddi, R, Valiyaveetil, F.I. | | Deposit date: | 2023-07-17 | | Release date: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | A facile approach for incorporating tyrosine esters to probe ion-binding sites and backbone hydrogen bonds.
J.Biol.Chem., 300, 2023
|
|
8THM
 
 | |
8TH9
 
 | |
8TH4
 
 | |
8TH3
 
 | |
8TH2
 
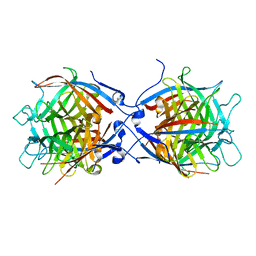 | | Structure of the isoflavene-forming dirigent protein PsPTS2 | | Descriptor: | Dirigent protein | | Authors: | Smith, C.A, Meng, Q, Lewis, N.G, Davin, L.B. | | Deposit date: | 2023-07-13 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Dirigent isoflavene-forming PsPTS2: 3D structure, stereochemical, and kinetic characterization comparison with pterocarpan-forming PsPTS1 homolog in pea.
J.Biol.Chem., 300, 2024
|
|
8TGH
 
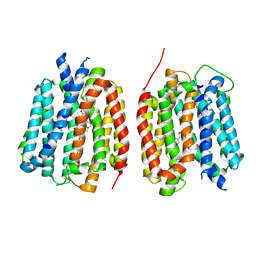 | | VMAT1 dimer with amphetamine and reserpine | | Descriptor: | (2S)-1-phenylpropan-2-amine, Chromaffin granule amine transporter, reserpine | | Authors: | Ye, J, Liu, B, Li, W. | | Deposit date: | 2023-07-12 | | Release date: | 2024-03-20 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural insights into vesicular monoamine storage and drug interactions.
Nature, 629, 2024
|
|
8TGC
 
 | |
8TGB
 
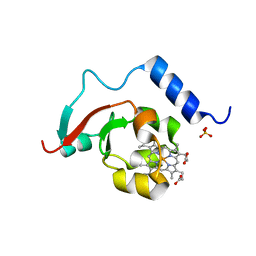 | | Crystal structure of root lateral formation protein (RLF) b5-domain from Oryza sativa | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SULFATE ION, root lateral formation protein (RLF) | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Benson, D.R. | | Deposit date: | 2023-07-12 | | Release date: | 2023-12-13 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | The N-terminal intrinsically disordered region of Ncb5or docks with the cytochrome b 5 core to form a helical motif that is of ancient origin.
Proteins, 92, 2024
|
|
8TG8
 
 | |
8TG7
 
 | |
