6GG0
 
 | |
8CSL
 
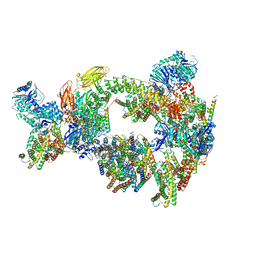 | | Sub-tomogram averaging of erythrocyte ankyrin-1 complex | | Descriptor: | Ammonium transporter Rh type A, Ankyrin-1, Band 3 anion transport protein, ... | | Authors: | Vallese, F, Kim, K, Yen, L.Y, Johnston, J.D, Noble, A.J, Cali, T, Clarke, O.B. | | Deposit date: | 2022-05-12 | | Release date: | 2022-07-20 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (25 Å) | | Cite: | Architecture of the human erythrocyte ankyrin-1 complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
6YP6
 
 | |
6GQG
 
 | |
6GQH
 
 | |
8D32
 
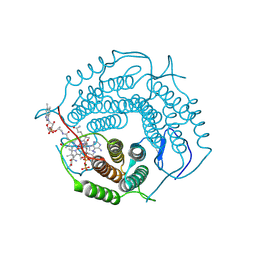 | | Mycobacterium tuberculosis pduO-type ATP:cobalamin adenosyltransferase bound to 5-deoxyadenosylrhodibalamin and PPPi | | Descriptor: | 5'-DEOXYADENOSINE, Corrinoid adenosyltransferase, MAGNESIUM ION, ... | | Authors: | Mascarenhas, R.N, Ruetz, M, Koutmos, M, Banerjee, R. | | Deposit date: | 2022-05-31 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Noble Metal Substitution Leads to B 12 Cofactor Mimicry by a Rhodibalamin.
Biochemistry, 63, 2024
|
|
6H1H
 
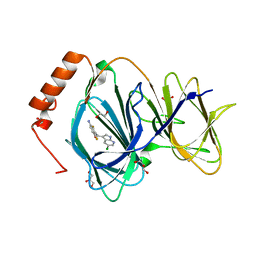 | | Crystal structure of human Pirin in complex with compound 7 (PLX4720) | | Descriptor: | 1,2-ETHANEDIOL, FE (III) ION, GLYCEROL, ... | | Authors: | Ali, S, Le Bihan, Y.V, van Montfort, R.L.M. | | Deposit date: | 2018-07-11 | | Release date: | 2018-11-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Privileged Structures and Polypharmacology within and between Protein Families.
ACS Med Chem Lett, 9, 2018
|
|
6HM1
 
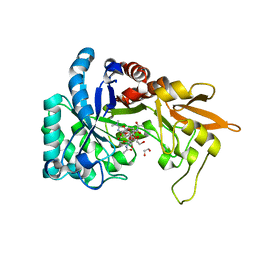 | | Structural and thermodynamic signatures of ligand binding to an enigmatic chitinase-D from Serratia proteamaculans | | Descriptor: | 1,2-ETHANEDIOL, ALLOSAMIDIN, Glycoside hydrolase family 18 | | Authors: | Madhuprakash, J, Dalhus, B, Vaaje-Kolstad, G, Eijsink, V.G.H, Sorlie, M. | | Deposit date: | 2018-09-11 | | Release date: | 2019-03-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Structural and Thermodynamic Signatures of Ligand Binding to the Enigmatic Chitinase D of Serratia proteamaculans.
J.Phys.Chem.B, 123, 2019
|
|
5E5X
 
 | |
8DJU
 
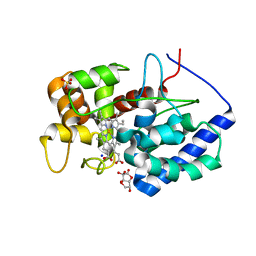 | | Cytosolic ascorbate peroxidase from Sorghum bicolor - bicyclic dehydroascorbic acid complex | | Descriptor: | (3aS,6S,6aR)-3,3,3a,6-tetrahydroxytetrahydrofuro[3,2-b]furan-2(3H)-one (non-preferred name), L-ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A sorghum ascorbate peroxidase with four binding sites has activity against ascorbate and phenylpropanoids.
Plant Physiol., 192, 2023
|
|
5KIZ
 
 | |
7QRM
 
 | | Cryo-EM structure of catalytically active Spinacia oleracea cytochrome b6f in complex with endogenous plastoquinones at 2.7 A resolution | | Descriptor: | (1S)-2-{[{[(2R)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL STEARATE, 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Sarewicz, M, Szwalec, M, Indyka, P, Rawski, M, Pintscher, S, Pietras, R, Mielecki, B, Jaciuk, M, Glatt, S, Osyczka, A. | | Deposit date: | 2022-01-11 | | Release date: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution cryo-EM structures of plant cytochrome b 6 f at work.
Sci Adv, 9, 2023
|
|
4X8C
 
 | | Crystal structure of human peptidylarginine deiminase type4 (PAD4) in complex with GSK147 | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type-4, [(3S,4R)-3-amino-4-hydroxypiperidin-1-yl]{2-[1-(cyclopropylmethyl)-1H-pyrrolo[2,3-b]pyridin-2-yl]-7-methoxy-1-methyl-1H-benzimidazol-5-yl}methanone | | Authors: | Lewis, H.D, Bax, B.D, Chung, C.-W, Polyakova, O, Thorpe, J. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation.
Nat.Chem.Biol., 11, 2015
|
|
4X8G
 
 | | Crystal structure of human peptidylarginine deiminase type4 (PAD4) in complex with GSK199 | | Descriptor: | CALCIUM ION, Protein-arginine deiminase type-4, [(3R)-3-aminopiperidin-1-yl][2-(1-ethyl-1H-pyrrolo[2,3-b]pyridin-2-yl)-7-methoxy-1-methyl-1H-benzimidazol-5-yl]methanone | | Authors: | Lewis, H.D, Bax, B.D, Chung, C.-W, Polyakova, O, Thorpe, J. | | Deposit date: | 2014-12-10 | | Release date: | 2015-01-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Inhibition of PAD4 activity is sufficient to disrupt mouse and human NET formation.
Nat.Chem.Biol., 11, 2015
|
|
8PKZ
 
 | |
8BVQ
 
 | | E. coli BAM complex (BamABCDE) bound to darobactin B | | Descriptor: | Darobactin-B, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, ... | | Authors: | Horne, J.E, Fenn, K.L, Radford, S.E, Ranson, N.A. | | Deposit date: | 2022-12-05 | | Release date: | 2023-05-24 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Darobactin B Stabilises a Lateral-Closed Conformation of the BAM Complex in E. coli Cells.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6OIK
 
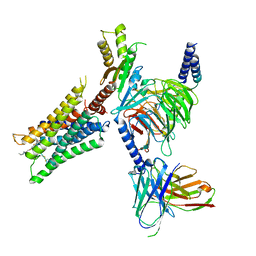 | | Muscarinic acetylcholine receptor 2-Go complex | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, 4-(4,5-dihydro-1,2-oxazol-3-yloxy)-N,N,N-trimethylbut-2-yn-1-aminium, Antibody fragment, ... | | Authors: | Maeda, S, Qianhui, Q, Skiniotis, G, Kobilka, B. | | Deposit date: | 2019-04-09 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structures of the M1 and M2 muscarinic acetylcholine receptor/G-protein complexes.
Science, 364, 2019
|
|
8TAD
 
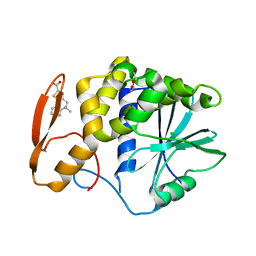 | | RTA in complex with inhibitor RUNT-206 | | Descriptor: | (9aM)-5,5-dimethyl-4,5-dihydronaphtho[1,2-b]thiophene-2-carboxylic acid, CHLORIDE ION, NONAETHYLENE GLYCOL, ... | | Authors: | Rudolph, M.J, Tumer, N. | | Deposit date: | 2023-06-27 | | Release date: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.76 Å) | | Cite: | Structure-based design and optimization of a new class of small molecule inhibitors targeting the P-stalk binding pocket of ricin.
Bioorg.Med.Chem., 100, 2024
|
|
4YTF
 
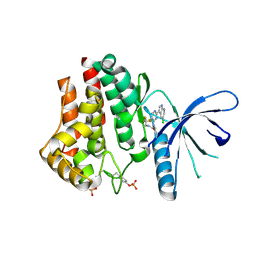 | | Discovery of VX-509 (Decernotinib): A Potent and Selective Janus kinase (JAK) 3 Inhibitor for the Treatment of Autoimmune Diseases | | Descriptor: | N~2~-[2-(5-chloro-1H-pyrrolo[2,3-b]pyridin-3-yl)-5-fluoropyrimidin-4-yl]-N-(2,2,2-trifluoroethyl)-L-alaninamide, Tyrosine-protein kinase JAK2 | | Authors: | Farmer, L, Ledeboer, M.W, Zuccola, H.J. | | Deposit date: | 2015-03-17 | | Release date: | 2015-08-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Discovery of VX-509 (Decernotinib): A Potent and Selective Janus Kinase 3 Inhibitor for the Treatment of Autoimmune Diseases.
J.Med.Chem., 58, 2015
|
|
8T9V
 
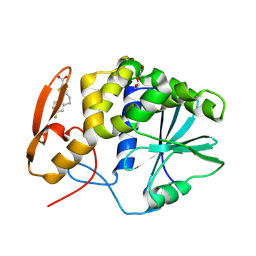 | | RTA-RUNT-59 complex structure | | Descriptor: | (9aP)-7-fluoro-4,5-dihydronaphtho[1,2-b]thiophene-2-carboxylic acid, NONAETHYLENE GLYCOL, Ricin | | Authors: | Rudolph, M.J, Tumer, N. | | Deposit date: | 2023-06-26 | | Release date: | 2024-05-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.945 Å) | | Cite: | Structure-based design and optimization of a new class of small molecule inhibitors targeting the P-stalk binding pocket of ricin.
Bioorg.Med.Chem., 100, 2024
|
|
7T96
 
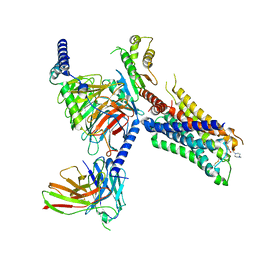 | | Cryo-EM structure of S2 state ACh-bound M2R-Go signaling complex with a PAM | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, ACETYLCHOLINE, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Xu, J, Wang, Q, Du, Y, Kobilka, B.K. | | Deposit date: | 2021-12-18 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.22 Å) | | Cite: | Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nat Commun, 14, 2023
|
|
7T94
 
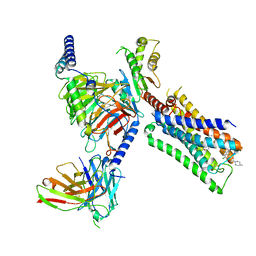 | | Cryo-EM structure of S1 state ACh-bound M2R-Go signaling complex with a PAM | | Descriptor: | 3-amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno[2,3-b]pyridine-2-carboxamide, ACETYLCHOLINE, Antibody fragment, ... | | Authors: | Xu, J, Wang, Q, Du, Y, Kobilka, B.K. | | Deposit date: | 2021-12-17 | | Release date: | 2023-01-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Structural and dynamic insights into supra-physiological activation and allosteric modulation of a muscarinic acetylcholine receptor.
Nat Commun, 14, 2023
|
|
4YTI
 
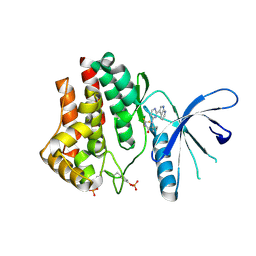 | |
7TEU
 
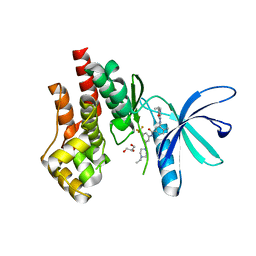 | | Crystal structure of JAK2 JH1 with type II inhibitor YLIU-04-105-1 | | Descriptor: | 3-{(4S)-2-[(cyclopropanecarbonyl)amino]imidazo[1,2-b]pyridazin-6-yl}-N-{3-[(4-ethylpiperazin-1-yl)methyl]-5-(trifluoromethyl)phenyl}-4-methylbenzamide, GLYCEROL, Tyrosine-protein kinase JAK2 | | Authors: | Hubbard, S.R. | | Deposit date: | 2022-01-05 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | New scaffolds for type II JAK2 inhibitors overcome the acquired G993A resistance mutation.
Cell Chem Biol, 30, 2023
|
|
8UM3
 
 | | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 6-chlorotetrazolo[1,5-b]pyridazine, ... | | Authors: | Godoy, A.S, Noske, G.D, Fairhead, M, Lithgo, R.M, Koekemoer, L, Aschenbrenner, J.C, Balcomb, B.H, Marples, P.G, Ni, X, Tomlinson, C.W.E, Wild, C, Mesquita, N.C.M.R, Oliva, G, Fearon, D, Walsh, M.A, von Delft, F. | | Deposit date: | 2023-10-17 | | Release date: | 2023-11-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.925 Å) | | Cite: | PanDDA analysis -- Crystal Structure of Zika virus NS3 Helicase in complex with Z203039992
To Be Published
|
|
