2PB1
 
 | | Exo-B-(1,3)-Glucanase from Candida Albicans in complex with unhydrolysed and covalently linked 2,4-dinitrophenyl-2-deoxy-2-fluoro-B-D-glucopyranoside at 1.9 A | | Descriptor: | 2,4-dinitrophenyl 2-deoxy-2-fluoro-beta-D-glucopyranoside, 2-deoxy-2-fluoro-alpha-D-glucopyranose, Hypothetical protein XOG1 | | Authors: | Cutfield, S.M, Cutfield, J.F, Davies, G.J, Sullivan, P.A. | | Deposit date: | 2007-03-28 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Exo-B-(1,3)-Glucanase from Candida Albicans in complex with unhydrolysed and covalently linked 2,4-dinitrophenyl-2-deoxy-2-fluoro-B-D-glucopyranoside at 1.9 A
To be Published
|
|
2PWH
 
 | | Crystal structure of the trehalulose synthase MutB from Pseudomonas mesoacidophila MX-45 | | Descriptor: | CALCIUM ION, Sucrose isomerase | | Authors: | Ravaud, S, Robert, X, Haser, R, Aghajari, N. | | Deposit date: | 2007-05-11 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Trehalulose synthase native and carbohydrate complexed structures provide insights into sucrose isomerization.
J.Biol.Chem., 61, 2007
|
|
1XOT
 
 | | Catalytic Domain Of Human Phosphodiesterase 4B In Complex With Vardenafil | | Descriptor: | 2-{2-ETHOXY-5-[(4-ETHYLPIPERAZIN-1-YL)SULFONYL]PHENYL}-5-METHYL-7-PROPYLIMIDAZO[5,1-F][1,2,4]TRIAZIN-4(1H)-ONE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-06 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
2NC9
 
 | | Apo solution structure of Hop TPR2A | | Descriptor: | Stress-induced-phosphoprotein 1 | | Authors: | Darby, J.F, Vidler, L.R, Simpson, P.J, Matthews, S.J, Sharp, S.Y, Pearl, L.H, Hoelder, S, Workman, P. | | Deposit date: | 2016-03-23 | | Release date: | 2017-03-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Hop TPR2A domain and investigation of target druggability by NMR, biochemical and in silico approaches.
Sci Rep, 10, 2020
|
|
1XMU
 
 | | Catalytic Domain Of Human Phosphodiesterase 4B In Complex With Roflumilast | | Descriptor: | 3-(CYCLOPROPYLMETHOXY)-N-(3,5-DICHLOROPYRIDIN-4-YL)-4-(DIFLUOROMETHOXY)BENZAMIDE, MAGNESIUM ION, ZINC ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-04 | | Release date: | 2004-12-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
1XOQ
 
 | | Catalytic Domain Of Human Phosphodiesterase 4D In Complex With Roflumilast | | Descriptor: | 1,2-ETHANEDIOL, 3-(CYCLOPROPYLMETHOXY)-N-(3,5-DICHLOROPYRIDIN-4-YL)-4-(DIFLUOROMETHOXY)BENZAMIDE, MAGNESIUM ION, ... | | Authors: | Card, G.L, England, B.P, Suzuki, Y, Fong, D, Powell, B, Lee, B, Luu, C, Tabrizizad, M, Gillette, S, Ibrahim, P.N, Artis, D.R, Bollag, G, Milburn, M.V, Kim, S.-H, Schlessinger, J, Zhang, K.Y.J. | | Deposit date: | 2004-10-06 | | Release date: | 2004-12-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Basis for the Activity of Drugs that Inhibit Phosphodiesterases.
STRUCTURE, 12, 2004
|
|
2NDB
 
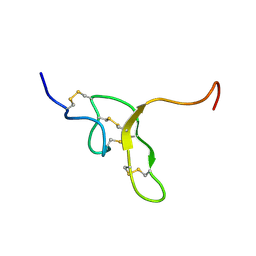 | |
2MRF
 
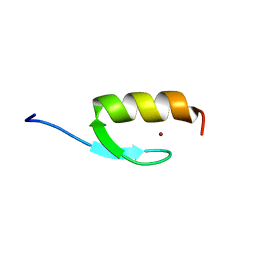 | | NMR structure of the ubiquitin-binding zinc finger (UBZ) domain from human Rad18 | | Descriptor: | E3 ubiquitin-protein ligase RAD18, ZINC ION | | Authors: | Rizzo, A.A, Salerno, P.E, Bezsonova, I, Korzhnev, D.M. | | Deposit date: | 2014-07-03 | | Release date: | 2014-10-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | NMR Structure of the Human Rad18 Zinc Finger in Complex with Ubiquitin Defines a Class of UBZ Domains in Proteins Linked to the DNA Damage Response.
Biochemistry, 53, 2014
|
|
2ANJ
 
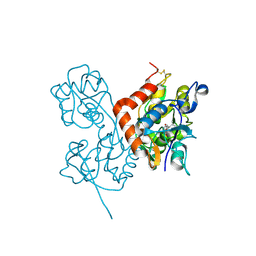 | | Crystal Structure of the Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant in Complex With the Partial Agonist Kainic Acid at 2.1 A Resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Holm, M.M, Naur, P, Vestergaard, B, Geballe, M.T, Gajhede, M, Kastrup, J.S, Traynelis, S.F, Egebjerg, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Binding Site Tyrosine Shapes Desensitization Kinetics and Agonist Potency at GluR2: a mutagenic, kinetic, and crystallographic study
J.Biol.Chem., 280, 2005
|
|
7MKO
 
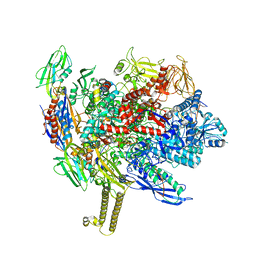 | | Escherichia coli RNA polymerase elongation complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural basis of RNA polymerase recycling by the Swi2/Snf2 family of ATPase RapA in Escherichia coli.
J.Biol.Chem., 297, 2021
|
|
7MKN
 
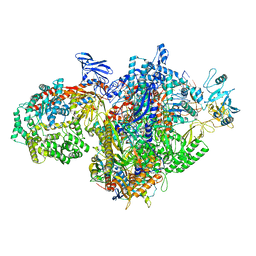 | | Escherichia coli RNA polymerase and RapA elongation complex | | Descriptor: | 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, DNA (29-MER), DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural basis of RNA polymerase recycling by the Swi2/Snf2 family of ATPase RapA in Escherichia coli.
J.Biol.Chem., 297, 2021
|
|
7MKQ
 
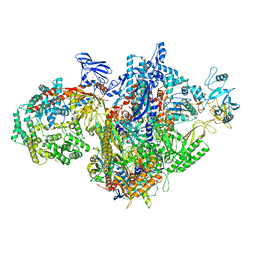 | | Escherichia coli RNA polymerase and RapA binary complex | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-02 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Structural basis of RNA polymerase recycling by the Swi2/Snf2 family of ATPase RapA in Escherichia coli.
J.Biol.Chem., 297, 2021
|
|
7MKP
 
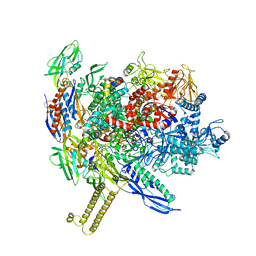 | | Escherichia coli RNA polymerase core enzyme | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Qayyum, M.Z, Murakami, K.S. | | Deposit date: | 2021-04-26 | | Release date: | 2021-06-23 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structural basis of RNA polymerase recycling by the Swi2/Snf2 family of ATPase RapA in Escherichia coli.
J.Biol.Chem., 297, 2021
|
|
7M5M
 
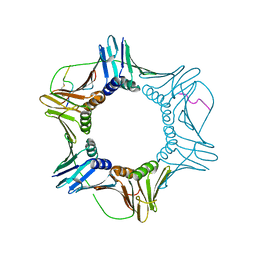 | | PCNA bound to peptide mimetic | | Descriptor: | N-BUTANE, Peptide mimetic (ACE)RQCSMTCFYHSK(NH2) with linker, Proliferating cell nuclear antigen | | Authors: | Vandborg, B.A, Bruning, J.B. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A cell permeable bimane-constrained PCNA-interacting peptide.
Rsc Chem Biol, 2, 2021
|
|
7M5L
 
 | | PCNA bound to peptide mimetic with linker | | Descriptor: | PROPANE, Peptide mimetic (ACE)RQCSMTCFYHSK(NH2) with linker, Proliferating cell nuclear antigen | | Authors: | Vandborg, B.A, Bruning, J.B. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | A cell permeable bimane-constrained PCNA-interacting peptide.
Rsc Chem Biol, 2, 2021
|
|
7M5N
 
 | | PCNA bound to peptide mimetic with linker | | Descriptor: | 1,3-dimethylbenzene, Peptide mimetic (ACE)RQCSMTCFYHSK(NH2) with linker, Proliferating cell nuclear antigen | | Authors: | Vandborg, B.A, Bruning, J.B. | | Deposit date: | 2021-03-24 | | Release date: | 2021-08-11 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | A cell permeable bimane-constrained PCNA-interacting peptide.
Rsc Chem Biol, 2, 2021
|
|
7NOY
 
 | | Crystal structure of the heterocyclic toxin methyltransferase from Mycobacterium tuberculosis in complex with substrate 1-hydroxyquinolin-4(1H)-one | | Descriptor: | 1-oxidanylquinolin-4-one, S-ADENOSYL-L-HOMOCYSTEINE, SODIUM ION, ... | | Authors: | Denkhaus, L, Sartor, P, Einsle, O, Gerhardt, S, Fetzner, S. | | Deposit date: | 2021-02-26 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of O-methylation of (2-heptyl-)1-hydroxyquinolin-4(1H)-one and related compounds by the heterocyclic toxin methyltransferase Rv0560c of Mycobacterium tuberculosis.
J.Struct.Biol., 213, 2021
|
|
7NMK
 
 | | Crystal structure of the heterocyclic toxin methyltransferase from Mycobacterium tuberculosis with bound methylation product 1-methoxyquinolin-4(1H)-one | | Descriptor: | 1-methoxy-4-oxoquinoline, 2-heptyl-1-hydroxyquinolin-4(1H)-one methyltransferase, FORMIC ACID, ... | | Authors: | Denkhaus, L, Sartor, P, Einsle, O, Gerhardt, S, Fetzner, S. | | Deposit date: | 2021-02-23 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.204 Å) | | Cite: | Structural basis of O-methylation of (2-heptyl-)1-hydroxyquinolin-4(1H)-one and related compounds by the heterocyclic toxin methyltransferase Rv0560c of Mycobacterium tuberculosis.
J.Struct.Biol., 213, 2021
|
|
7NDM
 
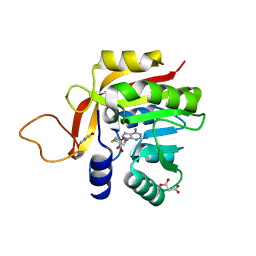 | | Crystal structure of the heterocyclic toxin methyltransferase from Mycobacterium tuberculosis with bound substrate 4-hydroxyisoquinolin-1(2H)-one | | Descriptor: | 4-oxidanyl-2~{H}-isoquinolin-1-one, Heterocyclic toxin methyltransferase (Rv0560c), MALONATE ION, ... | | Authors: | Denkhaus, L, Sartor, P, Einsle, O, Gerhardt, S, Fetzner, S. | | Deposit date: | 2021-02-02 | | Release date: | 2021-09-22 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural basis of O-methylation of (2-heptyl-)1-hydroxyquinolin-4(1H)-one and related compounds by the heterocyclic toxin methyltransferase Rv0560c of Mycobacterium tuberculosis.
J.Struct.Biol., 213, 2021
|
|
5MEE
 
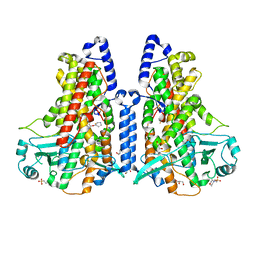 | | Cyanothece lipoxygenase 2 (CspLOX2) variant - L304V | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Arachidonate 15-lipoxygenase, FE (III) ION, ... | | Authors: | Newie, J, Neumann, P, Werner, M, Mata, R.A, Ficner, R, Feussner, I. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.
Sci Rep, 7, 2017
|
|
5MEG
 
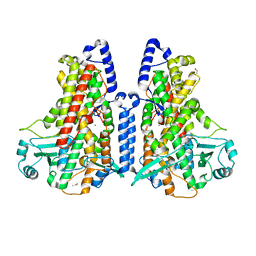 | | Manganese-substituted Cyanothece lipoxygenase 2 (Mn-CspLOX2) | | Descriptor: | Arachidonate 15-lipoxygenase, CHLORIDE ION, ETHANOL, ... | | Authors: | Newie, J, Neumann, P, Werner, M, Mata, R.A, Ficner, R, Feussner, I. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.
Sci Rep, 7, 2017
|
|
4M80
 
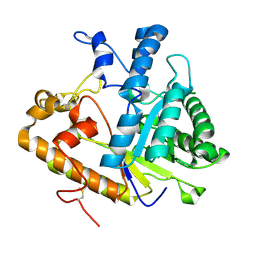 | | The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans at 1.85A resolution | | Descriptor: | EXO-1,3-BETA-GLUCANASE | | Authors: | Nakatani, Y, Cutfield, S.M, Larsen, D.S, Cutfield, J.F. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.858 Å) | | Cite: | Major Change in Regiospecificity for the Exo-1,3-beta-glucanase from Candida albicans following Its Conversion to a Glycosynthase.
Biochemistry, 53, 2014
|
|
5MEF
 
 | | Cyanothece lipoxygenase 2 (CspLOX2) variant - L304F | | Descriptor: | Arachidonate 15-lipoxygenase, CHLORIDE ION, FE (III) ION, ... | | Authors: | Newie, J, Neumann, P, Werner, M, Mata, R.A, Ficner, R, Feussner, I. | | Deposit date: | 2016-11-14 | | Release date: | 2017-05-31 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.357 Å) | | Cite: | Lipoxygenase 2 from Cyanothece sp. controls dioxygen insertion by steric shielding and substrate fixation.
Sci Rep, 7, 2017
|
|
4M81
 
 | | The structure of E292S glycosynthase variant of exo-1,3-beta-glucanase from Candida albicans complexed with 1-fluoro-alpha-D-glucopyranoside (donor) and p-nitrophenyl beta-D-glucopyranoside (acceptor) at 1.86A resolution | | Descriptor: | 4-nitrophenyl beta-D-glucopyranoside, EXO-1,3-BETA-GLUCANASE, GLYCEROL, ... | | Authors: | Nakatani, Y, Cutfield, S.M, Larsen, D.S, Cutfield, J.F. | | Deposit date: | 2013-08-12 | | Release date: | 2014-06-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Major Change in Regiospecificity for the Exo-1,3-beta-glucanase from Candida albicans following Its Conversion to a Glycosynthase.
Biochemistry, 53, 2014
|
|
7NV1
 
 | | Human Pol Kappa holoenzyme with Ub-PCNA | | Descriptor: | DNA Primer, DNA Template, DNA polymerase kappa, ... | | Authors: | Lancey, C, De Biasio, A, Hamdan, S.M. | | Deposit date: | 2021-03-15 | | Release date: | 2021-11-03 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (6.4 Å) | | Cite: | Cryo-EM structure of human Pol kappa bound to DNA and mono-ubiquitylated PCNA.
Nat Commun, 12, 2021
|
|
