3EEY
 
 | | CRYSTAL STRUCTURE OF PUTATIVE RRNA-METHYLASE FROM Clostridium thermocellum | | Descriptor: | GLYCEROL, Putative rRNA methylase, S-ADENOSYLMETHIONINE, ... | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Rutter, M, Hu, S, Bain, K, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-06 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of Rrna-Methylase from Clostridium Thermocellum
To be Published
|
|
2RRC
 
 | | Solution Structure of RNA aptamer against AML1 Runt domain | | Descriptor: | 5'-R(P*GP*GP*AP*CP*CP*CP*(AP7)P*CP*CP*AP*CP*GP*GP*CP*GP*AP*GP*GP*UP*CP*CP*A)-3' | | Authors: | Nomura, Y, Fujiwara, K, Chiba, M, Fukunaga, J, Tanaka, Y, Iibuchi, H, Tanaka, T, Nakamura, Y, Kawai, G, Kozu, T, Sakamoto, T. | | Deposit date: | 2010-06-23 | | Release date: | 2011-06-29 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A novel high affinity RNA motif that mimics DNA in AML1 Runt domain binding
To be Published
|
|
4Y7T
 
 | | Structural analysis of MurU | | Descriptor: | GLYCEROL, Nucleotidyl transferase, SULFATE ION | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
3CP2
 
 | | Crystal structure of GidA from E. coli | | Descriptor: | SULFATE ION, tRNA uridine 5-carboxymethylaminomethyl modification enzyme gidA | | Authors: | Scrima, A, Meyer, S, Versees, W, Wittinghofer, A. | | Deposit date: | 2008-03-30 | | Release date: | 2008-06-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structures of the conserved tRNA-modifying enzyme GidA: implications for its interaction with MnmE and substrate
J.Mol.Biol., 380, 2008
|
|
4Y7U
 
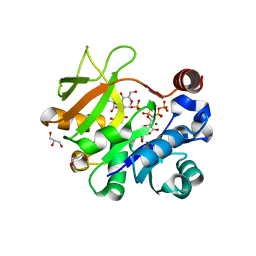 | | Structural analysis of MurU | | Descriptor: | 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]amino}phosphoryl]uridine, GLYCEROL, ... | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
4Y7V
 
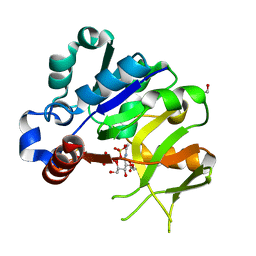 | | Structural analysis of MurU | | Descriptor: | 2-acetamido-3-O-[(1R)-1-carboxyethyl]-2-deoxy-1-O-phosphono-alpha-D-glucopyranose, GLYCEROL, IMIDODIPHOSPHORIC ACID, ... | | Authors: | Renner-Schneck, M.G, Stehle, T. | | Deposit date: | 2015-02-16 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the N-Acetylmuramic Acid alpha-1-Phosphate (MurNAc-alpha 1-P) Uridylyltransferase MurU, a Minimal Sugar Nucleotidyltransferase and Potential Drug Target Enzyme in Gram-negative Pathogens.
J.Biol.Chem., 290, 2015
|
|
8IU7
 
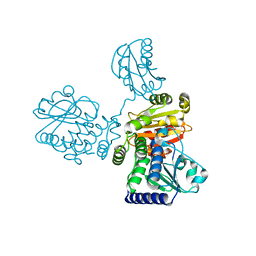 | |
3SGL
 
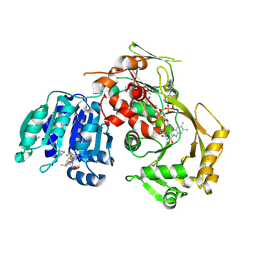 | |
4UZE
 
 | |
3FTM
 
 | |
2CFA
 
 | | Structure of viral flavin-dependant thymidylate synthase ThyX | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, THYMIDYLATE SYNTHASE | | Authors: | Graziani, S, Bernauer, J, Skouloubris, S, Graille, M, Zhou, C.-Z, Marchand, C, Decottignies, P, van Tilbeurgh, H, Myllykallio, H, Liebl, U. | | Deposit date: | 2006-02-17 | | Release date: | 2006-05-16 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Catalytic Mechanism and Structure of Viral Flavin-Dependent Thymidylate Synthase Thyx.
J.Biol.Chem., 281, 2006
|
|
3FS0
 
 | |
3E5Y
 
 | |
5WT2
 
 | | NifS from Helicobacter pylori | | Descriptor: | CHLORIDE ION, Cysteine desulfurase IscS, ISOPROPYL ALCOHOL, ... | | Authors: | Fujishiro, T, Takahashi, Y. | | Deposit date: | 2016-12-09 | | Release date: | 2017-12-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Snapshots of PLP-substrate and PLP-product external aldimines as intermediates in two types of cysteine desulfurase enzymes.
Febs J., 2019
|
|
1FSX
 
 | |
5OXS
 
 | |
7QCE
 
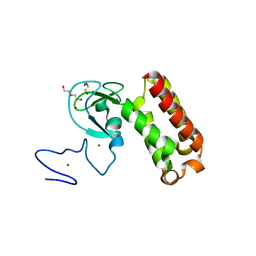 | | Crystal structure of an atypical PHD finger of VIN3 | | Descriptor: | DI(HYDROXYETHYL)ETHER, VIN3 (Protein VERNALIZATION INSENSITIVE 3), ZINC ION | | Authors: | Franco-Echevarria, E, Fiedler, M, Dean, C, Bienz, M. | | Deposit date: | 2021-11-23 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Plant vernalization proteins contain unusual PHD superdomains without histone H3 binding activity.
J.Biol.Chem., 298, 2022
|
|
2J3K
 
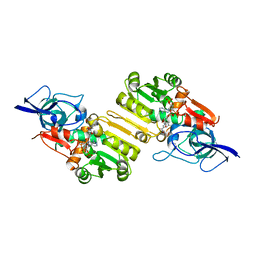 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex II | | Descriptor: | (2E,4R)-4-HYDROXYNON-2-ENAL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-22 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
2J3J
 
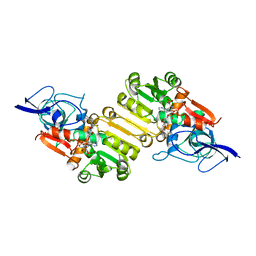 | | Crystal structure of Arabidopsis thaliana Double Bond Reductase (AT5G16970)-Ternary Complex I | | Descriptor: | 4'-HYDROXYCINNAMIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, NADPH-dependent oxidoreductase 2-alkenal reductase | | Authors: | Youn, B, Kim, S.J, Moinuddin, S.G, Lee, C, Bedgar, D.L, Harper, A.R, Davin, L.B, Lewis, N.G, Kang, C. | | Deposit date: | 2006-08-21 | | Release date: | 2006-10-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanistic and structural studies of apoform, binary, and ternary complexes of the Arabidopsis alkenal double bond reductase At5g16970.
J. Biol. Chem., 281, 2006
|
|
1SR5
 
 | | ANTITHROMBIN-ANHYDROTHROMBIN-HEPARIN TERNARY COMPLEX STRUCTURE | | Descriptor: | 2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-deoxy-2,3,6-tri-O-methyl-alpha-D-xylo-hexopyranose-(1-4)-2,3,6-tri-O-methyl-beta-D-glucopyranose-(1-4)-2,3-di-O-methyl-6-O-sulfo-alpha-D-glucopyranose-(1-4)-(2R,3R,4S,5S,6S)-6-(dihydroxymethyl)-3,4-dimethoxytetrahydro-2H-pyran-2,5-diol-(1-4)-2,3,6-tri-O-sulfo-alpha-D-glucopyranose-(1-4)-(2R,3R,4S,5S,6R)-6-(dihydroxymethyl)-3,4-dimethoxytetrahydro-2H-pyran-2,5-diol-(1-4)-1,5-anhydro-3-O-methyl-2,6-di-O-sulfo-D-glucitol, ... | | Authors: | Dementiev, A, Petitou, M, Gettins, P.G. | | Deposit date: | 2004-03-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | The ternary complex of antithrombin-anhydrothrombin-heparin reveals the basis of inhibitor specificity.
Nat.Struct.Mol.Biol., 11, 2004
|
|
1914
 
 | | SIGNAL RECOGNITION PARTICLE ALU RNA BINDING HETERODIMER, SRP9/14 | | Descriptor: | BETA-MERCAPTOETHANOL, PHOSPHATE ION, SIGNAL RECOGNITION PARTICLE 9/14 FUSION PROTEIN | | Authors: | Birse, D, Kapp, U, Strub, K, Cusack, S, Aberg, A. | | Deposit date: | 1997-11-13 | | Release date: | 1998-12-30 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | The crystal structure of the signal recognition particle Alu RNA binding heterodimer, SRP9/14.
EMBO J., 16, 1997
|
|
5OXR
 
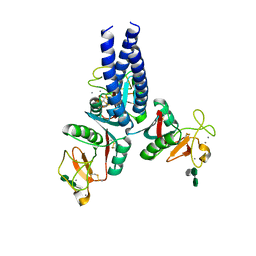 | |
1H8P
 
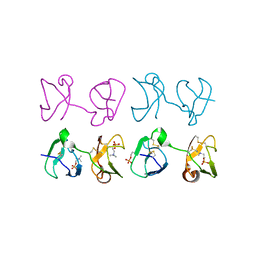 | | Bull seminal plasma PDC-109 fibronectin type II module | | Descriptor: | PHOSPHOCHOLINE, SEMINAL PLASMA PROTEIN PDC-109 | | Authors: | Wah, D.A, Fernandez-Tornero, C, Calvete, J.J, Romero, A. | | Deposit date: | 2001-02-14 | | Release date: | 2002-04-12 | | Last modified: | 2019-07-24 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Sperm Coating Mechanism from the 1.8 A Crystal Structure of Pdc-109-Phosphorylcholine Complex
Structure, 10, 2002
|
|
5F7U
 
 | |
5ZS9
 
 | |
