5KO6
 
 | | Crystal Structure of Isoform 2 of Purine Nucleoside Phosphorylase from Schistosoma mansoni in complex with cytosine and ribose-1-phosphate | | Descriptor: | 1-O-phosphono-alpha-D-ribofuranose, 6-AMINOPYRIMIDIN-2(1H)-ONE, Purine nucleoside phosphorylase | | Authors: | Torini, J.R, Romanello, L, Bird, L, Owens, R, Brandao-Neto, J, Pereira, H.M. | | Deposit date: | 2016-06-29 | | Release date: | 2017-08-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | The molecular structure of Schistosoma mansoni PNP isoform 2 provides insights into the nucleoside selectivity of PNPs.
PLoS ONE, 13, 2018
|
|
5KQ5
 
 | | AMPK bound to allosteric activator | | Descriptor: | 5'-AMP-activated protein kinase catalytic subunit alpha-1, 5'-AMP-activated protein kinase subunit beta-1, 5'-AMP-activated protein kinase subunit gamma-1, ... | | Authors: | Calabrese, M.F, Kurumbail, R.G. | | Deposit date: | 2016-07-05 | | Release date: | 2016-08-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Discovery and Preclinical Characterization of 6-Chloro-5-[4-(1-hydroxycyclobutyl)phenyl]-1H-indole-3-carboxylic Acid (PF-06409577), a Direct Activator of Adenosine Monophosphate-activated Protein Kinase (AMPK), for the Potential Treatment of Diabetic Nephropathy.
J.Med.Chem., 59, 2016
|
|
5HMV
 
 | | Re refinement of 4mwk. | | Descriptor: | CHLORIDE ION, DIMETHYL SULFOXIDE, Lysozyme C, ... | | Authors: | Helliwell, J.R. | | Deposit date: | 2016-01-17 | | Release date: | 2016-05-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (0.98 Å) | | Cite: | Comment on "Structural dynamics of cisplatin binding to histidine in a protein" [Struct. Dyn. 1, 034701 (2014)].
Struct Dyn, 3, 2016
|
|
5DYO
 
 | | Fab43.1 complex with flourescein | | Descriptor: | 2-(6-HYDROXY-3-OXO-3H-XANTHEN-9-YL)-BENZOIC ACID, Fab 43.1 Heavy Chain, Fab 43.1 Light Chain, ... | | Authors: | Longenecker, K.L, Judge, R.A. | | Deposit date: | 2015-09-25 | | Release date: | 2016-01-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Three-dimensional structure, binding, and spectroscopic characteristics of the monoclonal antibody 43.1 directed to the carboxyphenyl moiety of fluorescein.
Biopolymers, 105, 2016
|
|
5UIH
 
 | |
6STB
 
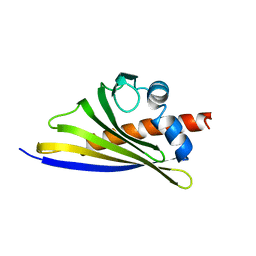 | | Crystal structure of the strawberry pathogenesis-related 10 (PR-10) Fra a 1.02 protein, Q64W mutant | | Descriptor: | Major strawberry allergen Fra a 1-2 | | Authors: | Orozco-Navarrete, B, Kaczmarska, Z, Dupeux, F, Pott, D, Diaz Perales, A, Casanal, A, Marquez, J.A, Valpuesta, V, Merchante, C. | | Deposit date: | 2019-09-10 | | Release date: | 2019-12-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structural Bases for the Allergenicity of Fra a 1.02 in Strawberry Fruits.
J.Agric.Food Chem., 68, 2020
|
|
5FM5
 
 | |
5ABD
 
 | | CRYSTAL STRUCTURE OF VEGFR-1 DOMAIN 2 IN PRESENCE OF CU | | Descriptor: | COPPER (II) ION, SODIUM ION, SULFATE ION, ... | | Authors: | Gaucher, J.-F, Lascombe, M.-B, Reille-Seroussi, M, Gagey-Eilstein, N, Broussy, S, Coric, P, Seijo, B, Gautier, B, Liu, W.-Q, Huguenot, F, Inguimbert, N, Bouaziz, S, Vidal, M, Broutin, I. | | Deposit date: | 2015-08-05 | | Release date: | 2016-09-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.995 Å) | | Cite: | Biophysical Studies of the Induced Dimerization of Human Vegf R Receptor 1 Binding Domain by Divalent Metals Competing with Vegf-A
Plos One, 11, 2016
|
|
5AFD
 
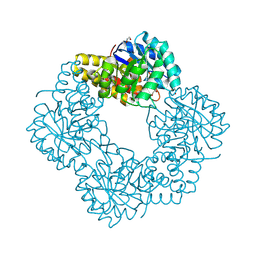 | | Native structure of N-acetylneuramininate lyase (sialic acid aldolase) from Aliivibrio salmonicida | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, N-ACETYLNEURAMINATE LYASE | | Authors: | Gurung, M.K, Altermark, B, Rader, I.L.U, Helland, R, Smalas, A.O. | | Deposit date: | 2015-01-21 | | Release date: | 2016-03-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Features and structure of a cold active N-acetylneuraminate lyase.
Plos One, 14, 2019
|
|
5T6E
 
 | |
1LO7
 
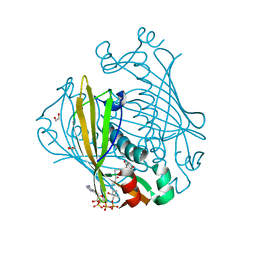 | | X-ray structure of 4-Hydroxybenzoyl CoA Thioesterase complexed with 4-hydroxyphenacyl CoA | | Descriptor: | 1,2-ETHANEDIOL, 4-HYDROXYPHENACYL COENZYME A, 4-hydroxybenzoyl-CoA Thioesterase | | Authors: | Thoden, J.B, Holden, H.M, Zhuang, Z, Dunaway-Mariano, D. | | Deposit date: | 2002-05-06 | | Release date: | 2002-05-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystallographic analyses of inhibitor and substrate complexes of wild-type and mutant 4-hydroxybenzoyl-CoA thioesterase.
J.Biol.Chem., 277, 2002
|
|
5T82
 
 | |
5ACC
 
 | | A Novel Oral Selective Estrogen Receptor Down-regulator, AZD9496, drives Tumour Growth Inhibition in Estrogen Receptor positive and ESR1 Mutant Models | | Descriptor: | (E)-3-(3,5-DIFLUORO-4-((1R,3R)-2-(2-FLUORO-2- METHYLPROPYL)-3-METHYL-2,3,4,9-TETRAHYDRO-1H-PYRIDO(3,4-B)INDOL-1-YL)PHENYL)ACRYLIC ACID, ESTROGEN RECEPTOR | | Authors: | Norman, R.A, Weir, H.M, Bradbury, R.H, Lawson, M, Rabow, A.A, Buttar, D, Callis, R.J, Curwen, J.O, de Almeida, C, Ballard, P, Hulse, M, Donald, C.S, Feron, L.J.L, Gingell, H, Karoutchi, G, MacFaul, P, Moss, T, Pearson, S.E, Tonge, M, Davies, G, Walker, G.E, Wilson, Z, Rowlinson, R, Powell, S, Hemsley, P, Linney, E, Campbell, H, Ghazoui, Z, Sadler, C, Richmond, G, Pazolli, E, Mazzola, A.M, DCruz, C, De Savi, C. | | Deposit date: | 2015-08-15 | | Release date: | 2015-12-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Optimization of a Novel Binding Motif to (E)-3-(3,5-Difluoro-4-((1R,3R)-2-(2-Fluoro-2-Methylpropyl)-3-Methyl-2, 3,4,9-Tetrahydro-1H-Pyrido[3,4-B]Indol-1-Yl)Phenyl)Acrylic Acid (Azd9496), a Potent and Orally Bioavailable Selective Estrogen Receptor Downregulator and Antagonist.
J.Med.Chem., 58, 2015
|
|
7BU6
 
 | | Structure of human beta1 adrenergic receptor bound to norepinephrine and nanobody 6B9 | | Descriptor: | (2S)-2,3-dihydroxypropyl octanoate, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, CHOLESTEROL, ... | | Authors: | Xu, X, Kaindl, J, Clark, M, Hubner, H, Hirata, K, Sunahara, R, Gmeiner, P, Kobilka, B.K, Liu, X. | | Deposit date: | 2020-04-04 | | Release date: | 2020-12-02 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Binding pathway determines norepinephrine selectivity for the human beta 1 AR over beta 2 AR.
Cell Res., 31, 2021
|
|
5D43
 
 | |
6U2J
 
 | | EM structure of MPEG-1 (L425K, alpha conformation) soluble pre-pore complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Macrophage-expressed gene 1 protein | | Authors: | Pang, S.S, Bayly-Jones, C. | | Deposit date: | 2019-08-20 | | Release date: | 2019-09-25 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The cryo-EM structure of the acid activatable pore-forming immune effector Macrophage-expressed gene 1.
Nat Commun, 10, 2019
|
|
5DIA
 
 | | PIM1 in complex with Cpd36 ((1S,3S)-N1-(6-(5-(pyridin-3-yl)-1H-pyrazolo[3,4-c]pyridin-3-yl)pyridin-2-yl)cyclohexane-1,3-diamine) | | Descriptor: | (1S,3S)-N-{6-[5-(pyridin-3-yl)-1H-pyrazolo[3,4-c]pyridin-3-yl]pyridin-2-yl}cyclohexane-1,3-diamine, Serine/threonine-protein kinase pim-1 | | Authors: | Murray, J.M, Wallweber, H. | | Deposit date: | 2015-08-31 | | Release date: | 2015-10-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.964 Å) | | Cite: | Discovery of 3,5-substituted 6-azaindazoles as potent pan-Pim inhibitors.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
7TN8
 
 | |
5FDI
 
 | | Crystal structure of Human Carbonic Anhydrase II with the anticonvulsant sulfamide JNJ-26990990 and its S,S-dioxide analog. | | Descriptor: | 1,1-bis(oxidanylidene)-3-[(sulfamoylamino)methyl]-1-benzothiophene, 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, ... | | Authors: | Di Fiore, A, De Simone, G, Alterio, V, Riccio, V, Winum, J.-Y, Carta, F, Supuran, C.T. | | Deposit date: | 2015-12-16 | | Release date: | 2016-05-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The anticonvulsant sulfamide JNJ-26990990 and its S,S-dioxide analog strongly inhibit carbonic anhydrases: solution and X-ray crystallographic studies.
Org.Biomol.Chem., 14, 2016
|
|
7EUW
 
 | | X-ray structure of high-strength hydrogel-grown FABP3 crystal soaked in 50% DMSO solution containing 4-[2-[1-(4-bromophenyl)-5-phenyl-1H-pyrazol-3-yl]phenoxy] (HA174) | | Descriptor: | 4-[2-[1-(4-bromophenyl)-5-phenyl-pyrazol-3-yl]phenoxy]butanoic acid, Fatty acid-binding protein, heart, ... | | Authors: | Sugiyama, S, Kakinouchi, K, Nakano, R, Matsuoka, S, Tsuchikawa, H, Sonoyama, M, Inoue, Y, Hayashi, F, Murata, M. | | Deposit date: | 2021-05-19 | | Release date: | 2022-05-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray structure of the human heart fatty acid-binding protein complexed with 4-[2-[1-(4-bromophenyl)-5-phenyl-1H-pyrazol-3-yl]phenoxy] (HA176)
To Be Published
|
|
6B72
 
 | | A novel HIV-1 Nef dimer interface induced by a single octyl-glucoside molecule | | Descriptor: | Protein Nef, octyl beta-D-glucopyranoside | | Authors: | Wu, M, Augelli-Szafran, C.E, Ptak, R.G, Smithgall, T.E. | | Deposit date: | 2017-10-03 | | Release date: | 2018-02-21 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A single beta-octyl glucoside molecule induces HIV-1 Nef dimer formation in the absence of partner protein binding.
PLoS ONE, 13, 2018
|
|
1JCL
 
 | | OBSERVATION OF COVALENT INTERMEDIATES IN AN ENZYME MECHANISM AT ATOMIC RESOLUTION | | Descriptor: | 1-HYDROXY-PENTANE-3,4-DIOL-5-PHOSPHATE, DEOXYRIBOSE-PHOSPHATE ALDOLASE | | Authors: | Heine, A, DeSantis, G, Luz, J.G, Mitchell, M, Wong, C.-H, Wilson, I.A. | | Deposit date: | 2001-06-09 | | Release date: | 2001-10-31 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | Observation of covalent intermediates in an enzyme mechanism at atomic resolution.
Science, 294, 2001
|
|
6TD2
 
 | |
5T1T
 
 | | Irak4 kinase - compound 1 co-structure | | Descriptor: | Interleukin-1 receptor-associated kinase 4, ~{N},~{N}-dimethyl-4-(6-nitroquinazolin-4-yl)oxy-cyclohexan-1-amine | | Authors: | Fischmann, T.O. | | Deposit date: | 2016-08-22 | | Release date: | 2017-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Identification of quinazoline based inhibitors of IRAK4 for the treatment of inflammation.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6B92
 
 | | Crystal Structure of the N-terminal domain of human METTL16 in complex with SAH | | Descriptor: | 1,2-ETHANEDIOL, S-ADENOSYL-L-HOMOCYSTEINE, U6 small nuclear RNA (adenine-(43)-N(6))-methyltransferase | | Authors: | Ruszkowska, A, Ruszkowski, M, Dauter, Z, Brown, J.A. | | Deposit date: | 2017-10-09 | | Release date: | 2018-04-04 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insights into the RNA methyltransferase domain of METTL16.
Sci Rep, 8, 2018
|
|
