4RFO
 
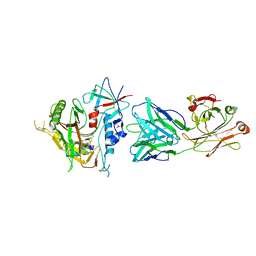 | | Crystal structure of the ADCC-Potent Antibody N60-I3 Fab in complex with HIV-1 Clade A/E gp120 and M48u1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HIV-1 clade A/E gp120, N60-i3 Fab heavy chain, ... | | Authors: | Tolbert, W.D, Gohain, N, Pazgier, M. | | Deposit date: | 2014-09-26 | | Release date: | 2015-07-15 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Cocrystal Structures of Antibody N60-i3 and Antibody JR4 in Complex with gp120 Define More Cluster A Epitopes Involved in Effective Antibody-Dependent Effector Function against HIV-1.
J.Virol., 89, 2015
|
|
4QYR
 
 | | Streptomyces platensis isomigrastatin ketosynthase domain MgsE KS3 | | Descriptor: | ACETIC ACID, AT-less polyketide synthase, CHLORIDE ION, ... | | Authors: | Kim, Y, Li, H, Endres, M, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-07-25 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
1JZN
 
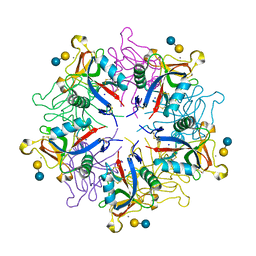 | | crystal structure of a galactose-specific C-type lectin | | Descriptor: | CALCIUM ION, CHLORIDE ION, Galactose-specific lectin, ... | | Authors: | Walker, J.R, Nagar, B, Young, N.M, Hirama, T, Rini, J.M. | | Deposit date: | 2001-09-16 | | Release date: | 2003-07-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | X-ray Crystal Structure of a Galactose-Specific C-Type Lectin Possessing a Novel Decameric Quaternary Structure.
Biochemistry, 43, 2004
|
|
2BAI
 
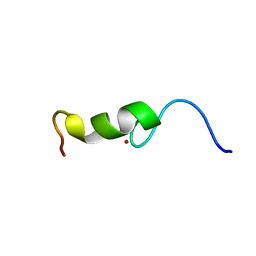 | | The Zinc finger domain of Mengovirus Leader polypeptide | | Descriptor: | Genome polyprotein, ZINC ION | | Authors: | Cornilescu, C.C, Porter, F.W, Qin, Z, Lee, M.S, Palmenberg, A.C, Markley, J.L, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2005-10-14 | | Release date: | 2006-01-24 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of the mengovirus Leader protein zinc-finger domain.
Febs Lett., 582, 2008
|
|
1O9S
 
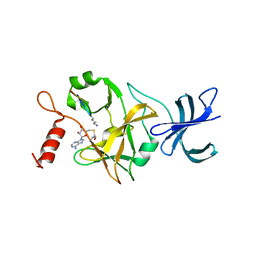 | | Crystal structure of a ternary complex of the human histone methyltransferase SET7/9 | | Descriptor: | GENE FRAGMENT FOR HISTONE H3, HISTONE-LYSINE N-METHYLTRANSFERASE, H3 LYSINE-4 SPECIFIC, ... | | Authors: | Xiao, B, Jing, C, Wilson, J.R, Walker, P.A, Vasisht, N, Kelly, G, Howell, S, Taylor, I.A, Blackburn, G.M, Gamblin, S.J. | | Deposit date: | 2002-12-18 | | Release date: | 2003-02-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Catalytic Mechanism of the Human Histone Methyltransferase Set7/9
Nature, 421, 2003
|
|
4JAR
 
 | |
4JAO
 
 | |
4JLY
 
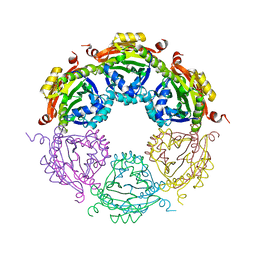 | | Dodecameric structure of spermidine N-acetyltransferase from Vibrio cholerae | | Descriptor: | SULFATE ION, Spermidine n1-acetyltransferase | | Authors: | Filippova, E.V, Minasov, G, Shuvalova, L, Kiryukhina, O, Kuhn, M.L, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-03-13 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.882 Å) | | Cite: | Substrate-Induced Allosteric Change in the Quaternary Structure of the Spermidine N-Acetyltransferase SpeG.
J.Mol.Biol., 427, 2015
|
|
2LW1
 
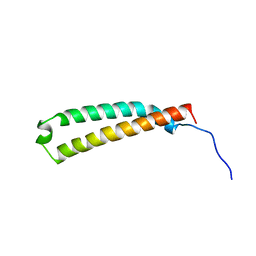 | | The C-terminal domain of the Uup protein is a DNA-binding coiled coil motif | | Descriptor: | ABC transporter ATP-binding protein uup | | Authors: | Carlier, L, Haase, A.S, Burgos Zepeda, M.Y, Dassa, E, Lequin, O. | | Deposit date: | 2012-07-19 | | Release date: | 2012-09-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | The C-terminal domain of the Uup protein is a DNA-binding coiled coil motif.
J.Struct.Biol., 180, 2012
|
|
2X0I
 
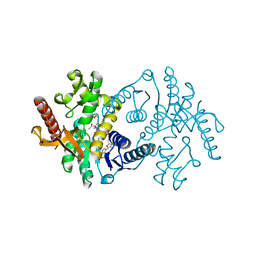 | | 2.9 A RESOLUTION STRUCTURE OF MALATE DEHYDROGENASE FROM ARCHAEOGLOBUS FULGIDUS IN COMPLEX WITH NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, MALATE DEHYDROGENASE, SODIUM ION, ... | | Authors: | Irimia, A, Madern, D, Zaccai, G, Vellieux, F.M.D, Karshikoff, A, Tibbelin, G, Ladenstein, R, Lien, T, Birkeland, N.-K. | | Deposit date: | 2009-12-14 | | Release date: | 2009-12-22 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | The 2.9A Resolution Crystal Structure of Malate Dehydrogenase from Archaeoglobus Fulgidus: Mechanisms of Oligomerisation and Thermal Stabilisation.
J.Mol.Biol., 335, 2004
|
|
4PF7
 
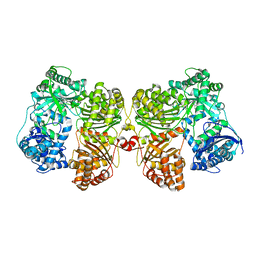 | | Crystal structure of insulin degrading enzyme complexed with inhibitor | | Descriptor: | (2S)-2-amino-N-{(1S)-1-cyclohexyl-2-[(4-methylphenyl)amino]-2-oxoethyl}-4-(methylselanyl)butanamide, Insulin-degrading enzyme, ZINC ION | | Authors: | Wang, Y, Guo, S. | | Deposit date: | 2014-04-28 | | Release date: | 2015-06-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Dual Exosite-binding Inhibitors of Insulin-degrading Enzyme Challenge Its Role as the Primary Mediator of Insulin Clearance in Vivo.
J.Biol.Chem., 290, 2015
|
|
4S35
 
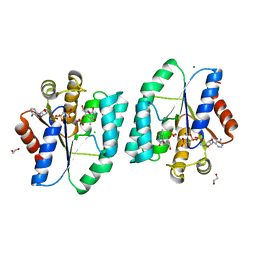 | | AMPPCP and TMP bound Crystal structure of thymidylate kinase (aq_969) from Aquifex Aeolicus VF5 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Biswas, A, Jeyakanthan, J, Sekar, K, Kuramitsu, S, Yokoyama, S. | | Deposit date: | 2015-01-26 | | Release date: | 2016-01-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural studies of a hyperthermophilic thymidylate kinase enzyme reveal conformational substates along the reaction coordinate.
Febs J., 284, 2017
|
|
2X9K
 
 | | Structure of a E.coli porin | | Descriptor: | OUTER MEMBRANE PROTEIN G, octyl beta-D-glucopyranoside | | Authors: | Korkmaz-Ozkan, F, Koster, S, Kuhlbrandt, W, Mantele, W, Yildiz, O. | | Deposit date: | 2010-03-21 | | Release date: | 2011-01-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Correlation between the Ompg Secondary Structure and its Ph-Dependent Alterations Monitored by Ftir.
J.Mol.Biol., 401, 2010
|
|
2XQ6
 
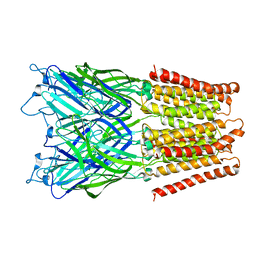 | | Pentameric ligand gated ion channel GLIC in complex with cesium ion (Cs+) | | Descriptor: | CESIUM ION, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
2XQA
 
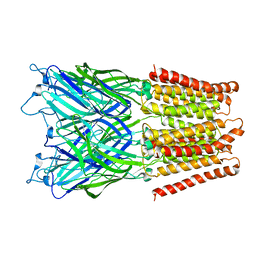 | | Pentameric ligand gated ion channel GLIC in complex with tetrabutylantimony (TBSb) | | Descriptor: | ANTIMONY (III) ION, GLR4197 PROTEIN | | Authors: | Hilf, R.J.C, Bertozzi, C, Zimmermann, I, Reiter, A, Trauner, D, Dutzler, R. | | Deposit date: | 2010-09-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural Basis of Open Channel Block in a Prokaryotic Pentameric Ligand-Gated Ion Channel
Nat.Struct.Mol.Biol., 17, 2010
|
|
3Q9N
 
 | | In silico and in vitro co-evolution of a high affinity complementary protein-protein interface | | Descriptor: | CARBAMOYL SARCOSINE, COENZYME A, CoA binding protein, ... | | Authors: | Karanicolas, J, Corn, J.E, Chen, I, Joachimiak, L.A, Dym, O, Chung, S, Albeck, S, Unger, T, Hu, W, Liu, G, Delbecq, S, Montelione, G.T, Spiegel, C, Liu, D, Baker, D, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2011-01-09 | | Release date: | 2011-04-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A de novo protein binding pair by computational design and directed evolution.
Mol.Cell, 42, 2011
|
|
4TSM
 
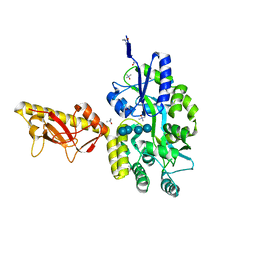 | | MBP-fusion protein of PilA1 from C. difficile R20291 residues 26-166 | | Descriptor: | alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, maltose-binding protein, pilin chimera, ... | | Authors: | Piepenbrink, K.H, Sundberg, E.J. | | Deposit date: | 2014-06-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Structural and Evolutionary Analyses Show Unique Stabilization Strategies in the Type IV Pili of Clostridium difficile.
Structure, 23, 2015
|
|
3QO8
 
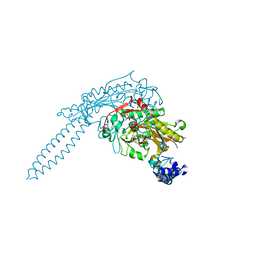 | | Crystal Structure of seryl-tRNA synthetase from Candida albicans | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, Seryl-tRNA synthetase, ... | | Authors: | Rocha, R, Santos, M.A, Pereira, P.J.B, Macedo-Ribeiro, S. | | Deposit date: | 2011-02-09 | | Release date: | 2011-08-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Unveiling the structural basis for translational ambiguity tolerance in a human fungal pathogen.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4PX2
 
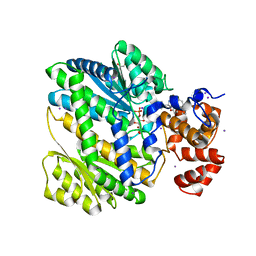 | | Human GKRP bound to AMG2882 and Sorbitol-6-Phosphate | | Descriptor: | D-SORBITOL-6-PHOSPHATE, GLYCEROL, Glucokinase regulatory protein, ... | | Authors: | Jordan, S.R, Chmait, S. | | Deposit date: | 2014-03-21 | | Release date: | 2015-05-13 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery and Structure Guided Optimization of Diarylmethanesulfonamide Disruptors of GK-GKRP Binding
To be Published
|
|
4Q0Q
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase in complex with L-ribulose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4Q0U
 
 | | Crystal structure of Acinetobacter sp. DL28 L-ribose isomerase mutant E204Q in complex with L-ribose | | Descriptor: | COBALT (II) ION, COBALT HEXAMMINE(III), L-Ribose isomerase, ... | | Authors: | Yoshida, H, Yoshihara, A, Teraoka, M, Izumori, K, Kamitori, S. | | Deposit date: | 2014-04-02 | | Release date: | 2014-05-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | X-ray structure of a novel L-ribose isomerase acting on a non-natural sugar L-ribose as its ideal substrate.
Febs J., 281, 2014
|
|
4PXS
 
 | |
4Q4E
 
 | | Crystal structure of E.coli aminopeptidase N in complex with actinonin | | Descriptor: | ACTINONIN, Aminopeptidase N, GLYCEROL, ... | | Authors: | Reddi, R, Ganji, R.J, Addlagatta, A. | | Deposit date: | 2014-04-14 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the inhibition of M1 family aminopeptidases by the natural product actinonin: Crystal structure in complex with E. coli aminopeptidase N.
Protein Sci., 24, 2015
|
|
4KKX
 
 | | Crystal structure of Tryptophan Synthase from Salmonella typhimurium with 2-aminophenol quinonoid in the beta site and the F6 inhibitor in the alpha site | | Descriptor: | 1,2-ETHANEDIOL, 2-{[4-(TRIFLUOROMETHOXY)BENZOYL]AMINO}ETHYL DIHYDROGEN PHOSPHATE, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hilario, E, Niks, D, Dunn, M.F, Mueller, L.J, Fan, L. | | Deposit date: | 2013-05-06 | | Release date: | 2014-01-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Allostery and substrate channeling in the tryptophan synthase bienzyme complex: evidence for two subunit conformations and four quaternary states.
Biochemistry, 52, 2013
|
|
2XSZ
 
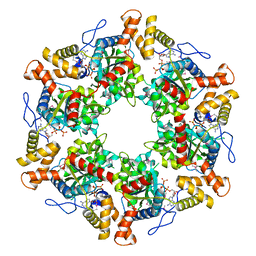 | | The dodecameric human RuvBL1:RuvBL2 complex with truncated domains II | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, RUVB-LIKE 1, RUVB-LIKE 2 | | Authors: | Gorynia, S, Bandeiras, T.M, Matias, P.M, Pinho, F.G, McVey, C.E, Vonrhein, C, Svergun, D.I, Round, A, Donner, P, Carrondo, M.A. | | Deposit date: | 2010-10-01 | | Release date: | 2011-10-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural and Functional Insights Into a Dodecameric Molecular Machine - the Ruvbl1/Ruvbl2 Complex.
J.Struct.Biol., 176, 2011
|
|
