7A89
 
 | |
8FHV
 
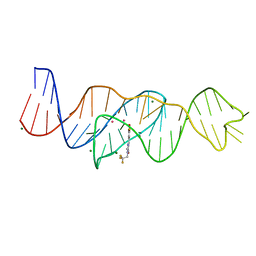 | | Structure of Lettuce aptamer bound to DFHBI-1T with thallium I ions | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-12-15 | | Release date: | 2023-05-10 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
1EME
 
 | |
4ZVP
 
 | |
4ZVS
 
 | |
4W7C
 
 | |
4ZVQ
 
 | |
4IK5
 
 | | High resolution structure of Delta-REST-GCaMP3 | | Descriptor: | CALCIUM ION, RCaMP, Green fluorescent protein | | Authors: | Chen, Y, Song, X, Miao, L, Zhu, Y, Ji, G. | | Deposit date: | 2012-12-25 | | Release date: | 2014-01-29 | | Last modified: | 2017-06-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insight into enhanced calcium indicator GCaMP3 and GCaMPJ to promote further improvement.
Protein Cell, 4, 2013
|
|
5MAK
 
 | | GFP-binding DARPin fusion gc_R7 | | Descriptor: | CITRIC ACID, Green fluorescent protein, R7 | | Authors: | Hansen, S, Stueber, J, Ernst, P, Koch, A, Bojar, D, Batyuk, A, Plueckthun, A. | | Deposit date: | 2016-11-03 | | Release date: | 2017-11-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Design and applications of a clamp for Green Fluorescent Protein with picomolar affinity.
Sci Rep, 7, 2017
|
|
7BYL
 
 | | Cryo-EM structure of human KCNQ4 | | Descriptor: | Calmodulin-3, Green fluorescent protein,Potassium voltage-gated channel subfamily KQT member 4, POTASSIUM ION, ... | | Authors: | Shen, H, Li, T, Yue, Z. | | Deposit date: | 2020-04-23 | | Release date: | 2020-12-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural Basis for the Modulation of Human KCNQ4 by Small-Molecule Drugs.
Mol.Cell, 81, 2021
|
|
1XA9
 
 | | Crystal structure of yellow fluorescent protein zFP538 K66M green mutant | | Descriptor: | BETA-MERCAPTOETHANOL, fluorescent protein FP538 | | Authors: | Remington, S.J, Wachter, R.M, Yarbrough, D.K, Branchaud, B, Anderson, D.C, Kallio, K, Lukyanov, K.A. | | Deposit date: | 2004-08-25 | | Release date: | 2005-02-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | zFP538, a yellow-fluorescent protein from Zoanthus, contains a novel three-ring chromophore.
Biochemistry, 44, 2005
|
|
6SXL
 
 | | Crystal structure of CrtE | | Descriptor: | Geranylgeranyl pyrophosphate synthase, PHOSPHATE ION | | Authors: | Feng, Y, Morgan, R.M.L, Nixon, P.J. | | Deposit date: | 2019-09-26 | | Release date: | 2020-06-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of Geranylgeranyl Pyrophosphate Synthase (CrtE) Involved in Cyanobacterial Terpenoid Biosynthesis.
Front Plant Sci, 11, 2020
|
|
1YFP
 
 | | STRUCTURE OF YELLOW-EMISSION VARIANT OF GFP | | Descriptor: | YELLOW FLUORESCENT PROTEIN | | Authors: | Wachter, R.M, Elsliger, M.-A, Kallio, K, Hanson, G.T, Remington, S.J. | | Deposit date: | 1998-08-28 | | Release date: | 1998-10-28 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of spectral shifts in the yellow-emission variants of green fluorescent protein.
Structure, 6, 1998
|
|
5GJP
 
 | |
6ZC5
 
 | | Human Adenovirus serotype D10 FiberKnob protein | | Descriptor: | Fiber | | Authors: | Baker, A.T, Mundy, R.M, Rizkallah, P.J, Parker, A.L. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Development of a low-seroprevalence, alpha v beta 6 integrin-selective virotherapy based on human adenovirus type 10.
Mol Ther Oncolytics, 25, 2022
|
|
8SCB
 
 | | Terminating ribosome with SRI-41315 | | Descriptor: | (2S,4aS)-2-cyclobutyl-10-methyl-3-phenyl-2,10-dihydropyrimido[4,5-b]quinoline-4,5(3H,4aH)-dione, 18S_rRNA, 28S_rRNA, ... | | Authors: | Yip, M.C.J, Coelho, J.P.L, Oltion, K, Tauton, J, Shao, S. | | Deposit date: | 2023-04-05 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | The eRF1 degrader SRI-41315 acts as a molecular glue at the ribosomal decoding center.
Nat.Chem.Biol., 20, 2024
|
|
7ZUX
 
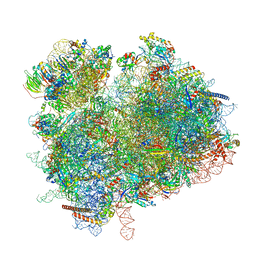 | | Collided ribosome in a disome unit from S. cerevisiae | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 40S ribosomal protein S0-A, ... | | Authors: | Best, K.M, Ikeuchi, K, Kater, L, Best, D.M, Musial, J, Matsuo, Y, Berninghausen, O, Becker, T, Inada, T, Beckmann, R. | | Deposit date: | 2022-05-13 | | Release date: | 2023-02-22 | | Last modified: | 2023-03-01 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Structural basis for clearing of ribosome collisions by the RQT complex.
Nat Commun, 14, 2023
|
|
7XNY
 
 | |
4X8H
 
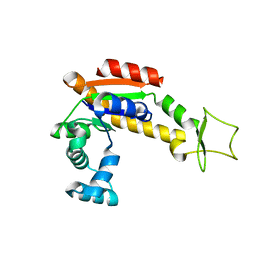 | | Crystal structure of E. coli Adenylate kinase P177A mutant | | Descriptor: | Adenylate kinase | | Authors: | Sauer-Eriksson, A.E, Kovermann, M, Aden, J, Grundstrom, C, Wolf-Watz, M, Sauer, U.H. | | Deposit date: | 2014-12-10 | | Release date: | 2015-07-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for catalytically restrictive dynamics of a high-energy enzyme state.
Nat Commun, 6, 2015
|
|
8JDJ
 
 | | Structure of the Human cytoplasmic Ribosome with human tRNA Asp(Q34) and mRNA(GAU) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Ishiguro, K, Yokoyama, T, Shirouzu, M, Suzuki, T. | | Deposit date: | 2023-05-14 | | Release date: | 2023-12-06 | | Last modified: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Glycosylated queuosines in tRNAs optimize translational rate and post-embryonic growth.
Cell, 186, 2023
|
|
1YHG
 
 | |
8FHX
 
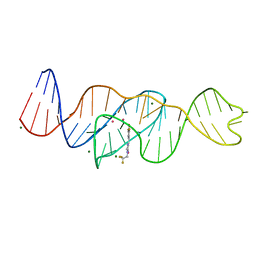 | | Structure of Lettuce aptamer bound to DFHBI-1T | | Descriptor: | (5Z)-5-(3,5-difluoro-4-hydroxybenzylidene)-2-methyl-3-(2,2,2-trifluoroethyl)-3,5-dihydro-4H-imidazol-4-one, Lettuce DNA aptamer, MAGNESIUM ION, ... | | Authors: | Passalacqua, L.F.M, Ferre-D'Amare, A.R. | | Deposit date: | 2022-12-15 | | Release date: | 2023-05-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Intricate 3D architecture of a DNA mimic of GFP.
Nature, 618, 2023
|
|
4RYW
 
 | |
5DPJ
 
 | | sfGFP double mutant - 133/149 p-ethynyl-L-phenylalanine | | Descriptor: | Green fluorescent protein | | Authors: | Dippel, A.B, Olenginski, G.M, Maurici, N, Liskov, M.T, Brewer, S.H, Phillips-Piro, C.M. | | Deposit date: | 2015-09-12 | | Release date: | 2016-02-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Probing the effectiveness of spectroscopic reporter unnatural amino acids: a structural study.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1HMY
 
 | |
