2GRY
 
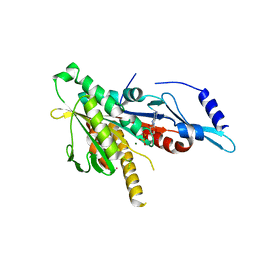 | | Crystal structure of the human KIF2 motor domain in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF2, MAGNESIUM ION, ... | | Authors: | Wang, J, Shen, Y, Tempel, W, Landry, R, Arrowsmith, C.H, Edwards, A.M, Sundstrom, M, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-04-25 | | Release date: | 2006-05-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the human KIF2 motor domain in complex with ADP
to be published
|
|
2PN5
 
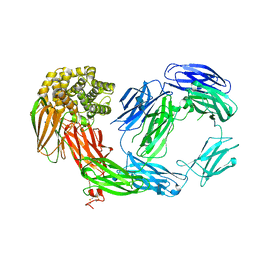 | | Crystal Structure of TEP1r | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SODIUM ION, ... | | Authors: | Baxter, R.H.G. | | Deposit date: | 2007-04-23 | | Release date: | 2007-07-24 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.698 Å) | | Cite: | Structural basis for conserved complement factor-like function in the antimalarial protein TEP1
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
2PHZ
 
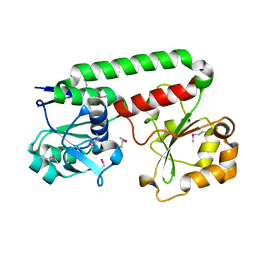 | | Crystal structure of Iron-uptake system-binding protein FeuA from Bacillus subtilis. Northeast Structural Genomics target SR580. | | Descriptor: | Iron-uptake system-binding protein | | Authors: | Benach, J, Neely, H, Seetharaman, J, Chen, C.X, Cunningham, K, Ma, L.-C, Janjua, H, Xiao, R, Baran, M, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-04-12 | | Release date: | 2007-04-24 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of Iron-uptake system-binding protein FeuA from Bacillus subtilis.
To be Published
|
|
3Q0H
 
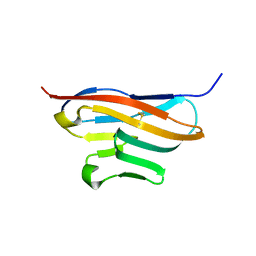 | | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT) | | Descriptor: | T cell immunoreceptor with Ig and ITIM domains | | Authors: | Ramagopal, U.A, Guo, H, Samanta, D, Nathenson, S.G, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2010-12-15 | | Release date: | 2011-02-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of T-cell immunoreceptor with immunoglobulin and ITIM domains (TIGIT)
To be published
|
|
3PJ5
 
 | |
2GXG
 
 | |
2PR9
 
 | | Mu2 adaptin subunit (AP50) of AP2 adaptor (second domain), complexed with GABAA receptor-gamma2 subunit-derived internalization peptide DEEYGYECL | | Descriptor: | AP-2 complex subunit mu-1, GABA(A) receptor subunit gamma-2 peptide | | Authors: | Vahedi-Faridi, A, Haucke, V, Kittler, J.T, Kukhtina, V, Moss, S.J, Saenger, W, Chen, G.-J, Tretter, V, Smith, K, Yan, Z, McAinsh, K, Arancibia-Carcamo, L. | | Deposit date: | 2007-05-04 | | Release date: | 2008-03-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Regulation of synaptic inhibition by phospho-dependent binding of the AP2 complex to a YECL motif in the GABAA receptor gamma2 subunit.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4KE8
 
 | | Crystal structure of Monoglyceride lipase from Bacillus sp. H257 in complex with monopalmitoyl glycerol analogue | | Descriptor: | Thermostable monoacylglycerol lipase, tetradecyl hydrogen (R)-(3-azidopropyl)phosphonate | | Authors: | Rengachari, S, Aschauer, P, Gruber, K, Dreveny, I, Oberer, M. | | Deposit date: | 2013-04-25 | | Release date: | 2013-09-18 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Conformational plasticity and ligand binding of bacterial monoacylglycerol lipase.
J.Biol.Chem., 288, 2013
|
|
3DHP
 
 | | Probing the role of aromatic residues at the secondary saccharide binding sites of human salivary alpha-amylase in substrate hydrolysis and bacterial binding | | Descriptor: | 4-amino-4,6-dideoxy-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, 5-HYDROXYMETHYL-CHONDURITOL, Alpha-amylase 1, ... | | Authors: | Ragunath, C, Manuel, S.G.A, Sait, H.M, Kasinathan, C. | | Deposit date: | 2008-06-18 | | Release date: | 2008-07-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Probing the role of aromatic residues
To be Published
|
|
3Q4I
 
 | | Crystal structure of CDP-Chase in complex with Gd3+ | | Descriptor: | GADOLINIUM ION, Phosphohydrolase (MutT/nudix family protein) | | Authors: | Duong-Ly, K.C, Gabelli, S.B, Amzel, L.M. | | Deposit date: | 2010-12-23 | | Release date: | 2011-07-13 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Nudix Hydrolase CDP-Chase, a CDP-Choline Pyrophosphatase, Is an Asymmetric Dimer with Two Distinct Enzymatic Activities.
J.Bacteriol., 193, 2011
|
|
3DBZ
 
 | | human surfactant protein D | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, Pulmonary surfactant-associated protein D | | Authors: | Head, J.F. | | Deposit date: | 2008-06-02 | | Release date: | 2008-08-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Interaction of recombinant surfactant protein D with lipopolysaccharide: conformation and orientation of bound protein by IRRAS and simulations.
Biochemistry, 47, 2008
|
|
2PS3
 
 | | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli | | Descriptor: | High-affinity zinc uptake system protein znuA | | Authors: | Yatsunyk, L.A, Kim, L.R, Vorontsov, I.I, Rosenzweig, A.C. | | Deposit date: | 2007-05-04 | | Release date: | 2007-06-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Structure and metal binding properties of ZnuA, a periplasmic zinc transporter from Escherichia coli.
J.Biol.Inorg.Chem., 13, 2008
|
|
4KF9
 
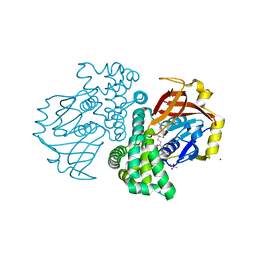 | | Crystal structure of a glutathione transferase family member from ralstonia solanacearum, target efi-501780, with bound gsh coordinated to a zinc ion, ordered active site | | Descriptor: | ACETATE ION, GLUTATHIONE, Glutathione s-transferase protein, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Armstrong, R.N, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-26 | | Release date: | 2013-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a glutathione transferase family member from ralstonia solanacearum, target efi-501780, with bound gsh coordinated to a zinc ion, ordered active site
To be Published
|
|
2PLY
 
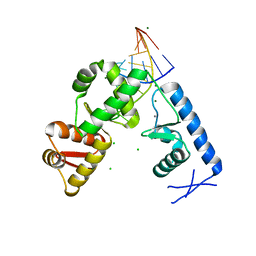 | | Structure of the mRNA binding fragment of elongation factor SelB in complex with SECIS RNA. | | Descriptor: | CALCIUM ION, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Soler, N, Fourmy, D, Yoshizawa, S. | | Deposit date: | 2007-04-20 | | Release date: | 2007-11-06 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural insight into a molecular switch in tandem winged-helix motifs from elongation factor SelB.
J.Mol.Biol., 370, 2007
|
|
3Q51
 
 | |
2PSS
 
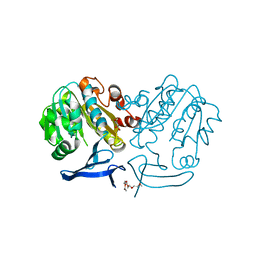 | | The structure of Plasmodium falciparum spermidine synthase in its apo-form | | Descriptor: | 2-(2-{2-[2-(2-METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHANOL, GLYCEROL, SULFATE ION, ... | | Authors: | Dufe, V.T, Qiu, W, Muller, I.B, Hui, R, Walter, R.D, Al-Karadaghi, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-05-07 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Plasmodium falciparum spermidine synthase in complex with the substrate decarboxylated S-adenosylmethionine and the potent inhibitors 4MCHA and AdoDATO.
J.Mol.Biol., 373, 2007
|
|
3DI8
 
 | | Crystal structure of bovine pancreatic ribonuclease A variant (V57A) | | Descriptor: | CHLORIDE ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Kurpiewska, K, Font, J, Ribo, M, Vilanova, M, Lewinski, K. | | Deposit date: | 2008-06-20 | | Release date: | 2008-07-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | X-ray crystallographic studies of RNase A variants engineered at the most destabilizing positions of the main hydrophobic core: further insight into protein stability
Proteins, 77, 2009
|
|
3DCK
 
 | | X-ray structure of D25N chemical analogue of HIV-1 protease complexed with ketomethylene isostere inhibitor | | Descriptor: | (2S)-2-{[(2R,5S)-5-{[(2S,3S)-2-{[(2S,3R)-2-(acetylamino)-3-hydroxybutanoyl]amino}-3-methylpentanoyl]amino}-2-butyl-4-oxononanoyl]amino}-N~1~-[(2S)-1-amino-5-carbamimidamido-1-oxopentan-2-yl]pentanediamide, Chemical analogue HIV-1 protease | | Authors: | Torbeev, V.Y, Mandal, K, Terechko, V.A, Kent, S.B.H. | | Deposit date: | 2008-06-03 | | Release date: | 2008-08-19 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of chemically synthesized HIV-1 protease and a ketomethylene isostere inhibitor based on the p2/NC cleavage site
Bioorg.Med.Chem.Lett., 18, 2008
|
|
2GVY
 
 | |
3DCR
 
 | | X-ray structure of HIV-1 protease and hydrated form of ketomethylene isostere inhibitor | | Descriptor: | Chemical analogue HIV-1 protease, N~2~-[(2R,5S)-5-({(2S,3S)-2-[(N-acetyl-L-threonyl)amino]-3-methylpent-4-enoyl}amino)-2-butyl-4,4-dihydroxynonanoyl]-L-glutaminyl-L-argininamide | | Authors: | Torbeev, V.Y, Mandal, K, Terechko, V.A, Kent, S.B.H. | | Deposit date: | 2008-06-04 | | Release date: | 2008-08-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of chemically synthesized HIV-1 protease and a ketomethylene isostere inhibitor based on the p2/NC cleavage site
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3PGY
 
 | | Serine hydroxymethyltransferase from Staphylococcus aureus, S95P mutant. | | Descriptor: | 1,2-ETHANEDIOL, CITRIC ACID, GLYCINE, ... | | Authors: | Osipiuk, J, Makowska-Grzyska, M, Kwon, K, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-11-02 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Serine hydroxymethyltransferase from Staphylococcus aureus, S95P mutant
To be Published
|
|
2EHW
 
 | |
4O94
 
 | | Crystal structure of a trap periplasmic solute binding protein from Rhodopseudomonas palustris HaA2 (RPB_3329), Target EFI-510223, with bound succinate | | Descriptor: | CHLORIDE ION, SUCCINIC ACID, TRAP dicarboxylate transporter DctP subunit | | Authors: | Vetting, M.W, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2014-01-01 | | Release date: | 2014-01-22 | | Last modified: | 2015-02-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
3PNL
 
 | | Crystal Structure of E.coli Dha kinase DhaK-DhaL complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Shi, R, McDonald, L, Matte, A, Cygler, M, Ekiel, I, Montreal-Kingston Bacterial Structural Genomics Initiative (BSGI) | | Deposit date: | 2010-11-19 | | Release date: | 2011-01-12 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
4KKN
 
 | | Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704 | | Descriptor: | Cytotoxic T-lymphocyte associated protein 4, alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kumar, P.R, Ahmed, M, Banu, R, Bhosle, R, Bonanno, J, Calarese, D.A, Celikgil, A, Chamala, S, Chan, M.K, Chowdhury, S, Fiser, A, Garforth, S.J, Scott Glenn, A, Hammonds, J, Hillerich, B, Khafizov, K, Lafleur, J, Attonito, J, Love, J.D, Patel, H, Patel, R, Seidel, R.D, Smith, B, Stead, M, Toro, R, Casadevall, A, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC), Atoms-to-Animals: The Immune Function Network (IFN) | | Deposit date: | 2013-05-06 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.253 Å) | | Cite: | Crystal structure of bovine CTLA-4, PSI-NYSGRC-012704
to be published
|
|
