3A07
 
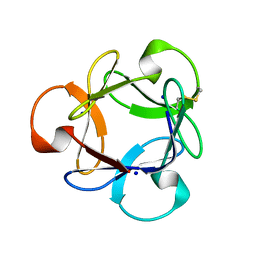 | | Crystal Structure of Actinohivin; Potent anti-HIV Protein | | Descriptor: | Actinohivin, SODIUM ION | | Authors: | Tsunoda, M, Suzuki, K, Sagara, T, Takenaka, A. | | Deposit date: | 2009-03-04 | | Release date: | 2009-08-25 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Mechanism by which the lectin actinohivin blocks HIV infection of target cells
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3A8W
 
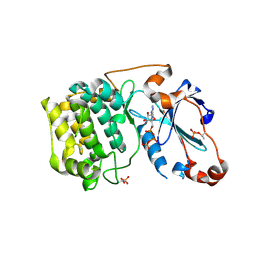 | | Crystal Structure of PKCiota kinase domain | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Protein kinase C iota type, SULFATE ION | | Authors: | Takimura, T, Kamata, K. | | Deposit date: | 2009-10-11 | | Release date: | 2010-05-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of the PKC-iota kinase domain in its ATP-bound and apo forms reveal defined structures of residues 533-551 in the C-terminal tail and their roles in ATP binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2ZXZ
 
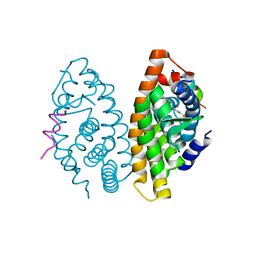 | | Crystal structure of the human RXR alpha ligand binding domain bound to a synthetic agonist compound and a coactivator peptide | | Descriptor: | 4-[2-(1,1,3,3-tetramethyl-2,3-dihydro-1H-inden-5-yl)-1,3-dioxolan-2-yl]benzoic acid, GRIP1 from Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Sato, Y, Antony, P, Rochel, N, Moras, D, Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Silicon analogues of the RXR-selective retinoid agonist SR11237 (BMS649): chemistry and biology
Chemmedchem, 4, 2009
|
|
2ZY0
 
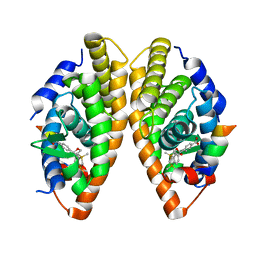 | | Crystal structure of the human RXR alpha ligand binding domain bound to a synthetic agonist compound and a coactivator peptide | | Descriptor: | 4-[2-(1,1,3,3-tetramethyl-2,3-dihydro-1H-1,3-benzodisilol-5-yl)-1,3-dioxolan-2-yl]benzoic acid, GRIP1 from Nuclear receptor coactivator 2, Retinoic acid receptor RXR-alpha | | Authors: | Sato, Y, Antony, P, Rochel, N, Moras, D, Structural Genomics Consortium for Research on Gene Expression (SGCGES) | | Deposit date: | 2009-01-09 | | Release date: | 2009-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Silicon analogues of the RXR-selective retinoid agonist SR11237 (BMS649): chemistry and biology
Chemmedchem, 4, 2009
|
|
3A9U
 
 | |
5MMT
 
 | |
3A3V
 
 | | Crystal structure of reducing-end-xylose releasing exo-oligoxylanase Y198F mutant | | Descriptor: | GLYCEROL, NICKEL (II) ION, Xylanase Y | | Authors: | Hidaka, M, Fushinobu, S, Honda, Y, Kitaoka, M. | | Deposit date: | 2009-06-22 | | Release date: | 2009-11-03 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Structural explanation for the acquisition of glycosynthase activity
J.Biochem., 147, 2010
|
|
3A8X
 
 | | Crystal Structure of PKCiota kinase domain | | Descriptor: | Protein kinase C iota type, SULFATE ION | | Authors: | Takimura, T, Kamata, K. | | Deposit date: | 2009-10-11 | | Release date: | 2010-05-05 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structures of the PKC-iota kinase domain in its ATP-bound and apo forms reveal defined structures of residues 533-551 in the C-terminal tail and their roles in ATP binding
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3A1L
 
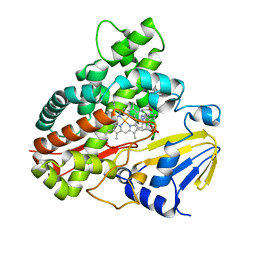 | | Crystal Structure of 11,11'-Dichlorochromopyrrolic Acid Bound Cytochrome P450 StaP (CYP245A1) | | Descriptor: | 3,4-bis(7-chloro-1H-indol-3-yl)-1H-pyrrole-2,5-dicarboxylic acid, Cytochrome P450, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Makino, M, Sugimoto, H, Shiro, Y, Asamizu, S, Onaka, H, Nagano, S. | | Deposit date: | 2009-04-09 | | Release date: | 2009-05-12 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Theoretical and experimental studies of the conversion of chromopyrrolic acid to an antitumor derivative by cytochrome P450 StaP: the catalytic role of water molecules
J.Am.Chem.Soc., 131, 2009
|
|
3A4Y
 
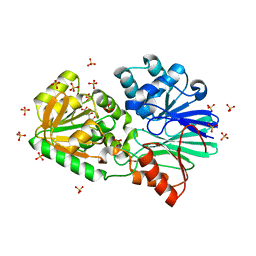 | | Crystal Structure of H61A mutant TTHA0252 from Thermus thermophilus HB8 | | Descriptor: | CITRATE ANION, Ribonuclease TTHA0252, SULFATE ION, ... | | Authors: | Ishikawa, H, Nakagawa, N, Kuramitsu, S, Yokoyama, S, Masui, R, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2009-07-22 | | Release date: | 2009-08-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal Structure of H61A mutant TTHA0252 from Thermus thermophilus HB8
to be published
|
|
2ZZE
 
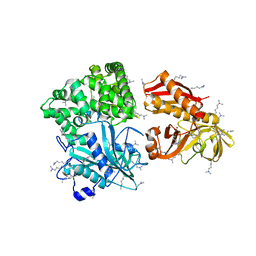 | | Crystal structure of alanyl-tRNA synthetase without oligomerization domain in lysine-methylated form | | Descriptor: | Alanyl-tRNA synthetase, ZINC ION | | Authors: | Sokabe, M, Ose, T, Tokunaga, K, Nakamura, A, Nureki, O, Yao, M, Tanaka, I. | | Deposit date: | 2009-02-10 | | Release date: | 2009-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | The structure of alanyl-tRNA synthetase with editing domain.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
3ACL
 
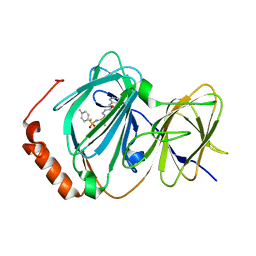 | | Crystal Structure of Human Pirin in complex with Triphenyl Compound | | Descriptor: | FE (II) ION, N-{[4-(benzyloxy)phenyl](methyl)-lambda~4~-sulfanylidene}-4-methylbenzenesulfonamide, Pirin | | Authors: | Okumura, H, Miyazaki, I, Simizu, S, Osada, H. | | Deposit date: | 2010-01-05 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | A small-molecule inhibitor shows that pirin regulates migration of melanoma cells
Nat.Chem.Biol., 6, 2010
|
|
3AFR
 
 | | Crystal Structure of VDR-LBD/22S-Butyl-1a,24R-dihydroxyvitamin D3 complex | | Descriptor: | (1R,3S,5Z)-5-[(2E)-2-{(1R,3aS,7aR)-1-[(1R,2S,4R)-2-butyl-4-hydroxy-1,5-dimethylhexyl]-7a-methyloctahydro-4H-inden-4-yli dene}ethylidene]-4-methylidenecyclohexane-1,3-diol, 13-meric peptide from Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Inaba, Y, Nakabayashi, M, Itoh, T, Ikura, T, Ito, N, Yamamoto, K. | | Deposit date: | 2010-03-10 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 22S-Butyl-1alpha,24R-dihydroxyvitamin D(3): Recovery of vitamin D receptor agonistic activity
J.Steroid Biochem.Mol.Biol., 121, 2010
|
|
3AL6
 
 | | Crystal structure of Human TYW5 | | Descriptor: | 2-OXOGLUTARIC ACID, JmjC domain-containing protein C2orf60, NICKEL (II) ION | | Authors: | Kato, M, Araiso, Y, Ishitani, R, Nureki, O. | | Deposit date: | 2010-07-26 | | Release date: | 2010-12-01 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a novel JmjC-domain-containing protein, TYW5, involved in tRNA modification.
Nucleic Acids Res., 39, 2011
|
|
3AS8
 
 | | MamA MSR-1 P41212 | | Descriptor: | Magnetosome protein MamA, SULFATE ION | | Authors: | Zeytuni, N, Davidov, G, Zarivach, R. | | Deposit date: | 2010-12-10 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ASH
 
 | | MamA D159K mutant 1 | | Descriptor: | MamA, SULFATE ION | | Authors: | Zeytuni, N, Levin, M, Davidov, G, Zarivach, R. | | Deposit date: | 2010-12-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3APF
 
 | | Crystal structure of human PI3K-gamma in complex with CH5039699 | | Descriptor: | 3-[7-(1H-benzimidazol-5-yl)-2-(morpholin-4-yl)-6,7-dihydro-5H-pyrrolo[2,3-d]pyrimidin-4-yl]phenol, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Nakamura, M, Fukami, T.A, Miyazaki, T, Yoshida, M. | | Deposit date: | 2010-10-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Discovery and biological activity of a novel class I PI3K inhibitor, CH5132799
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3ARP
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with DEQUALINIUM | | Descriptor: | Chitinase A, DEQUALINIUM, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3AS1
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - W275G mutant complex structure with chelerythrine | | Descriptor: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3AV5
 
 | | Crystal structure of mouse DNA methyltransferase 1 with AdoHcy | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AV6
 
 | | Crystal structure of mouse DNA methyltransferase 1 with AdoMet | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Takeshita, K, Suetake, I, Yamashita, E, Suga, M, Narita, H, Nakagawa, A, Tajima, S. | | Deposit date: | 2011-02-22 | | Release date: | 2011-05-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.09 Å) | | Cite: | Structural insight into maintenance methylation by mouse DNA methyltransferase 1 (Dnmt1).
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3ARW
 
 | | Crystal Structure Analysis of Chitinase A from Vibrio harveyi with novel inhibitors - complex structure with chelerythrine | | Descriptor: | 1,2-dimethoxy-12-methyl[1,3]benzodioxolo[5,6-c]phenanthridin-12-ium, Chitinase A, GLYCEROL | | Authors: | Pantoom, S, Vetter, I.R, Prinz, H, Suginta, W. | | Deposit date: | 2010-12-09 | | Release date: | 2011-04-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent family-18 chitinase inhibitors: x-ray structures, affinities, and binding mechanisms
J.Biol.Chem., 286, 2011
|
|
3ASG
 
 | | MamA D159K mutant 2 | | Descriptor: | MamA | | Authors: | Zeytuni, N, Levin, M, Davidov, G, Zarivach, R. | | Deposit date: | 2010-12-11 | | Release date: | 2011-07-20 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3AYH
 
 | |
3AWR
 
 | |
