4J9D
 
 | |
7Y4M
 
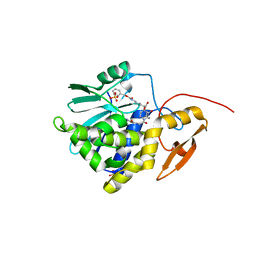 | | Crystal structure of Ricin A chain bound with N2-(2-amino-4-oxo-3,4-dihydropteridine-7-carbonyl)glycyl-L-phenylalanyl-N6-((benzyloxy)carbonyl)-L-lysine | | Descriptor: | (2S)-2-[[(2S)-2-[2-[(2-azanyl-4-oxidanylidene-3H-pteridin-7-yl)carbonylamino]ethanoylamino]-3-phenyl-propanoyl]amino]-6-(phenylmethoxycarbonylamino)hexanoic acid, Ricin A chain, SULFATE ION | | Authors: | Katakura, S, Goto, M, Ohba, T, Kawata, R, Nagatsu, K, Higashi, S, Matsumoto, K, Kurisu, K, Ohtsuka, K, Saito, R. | | Deposit date: | 2022-06-15 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Pterin-based small molecule inhibitor capable of binding to the secondary pocket in the active site of ricin-toxin A chain.
Plos One, 17, 2022
|
|
4J9B
 
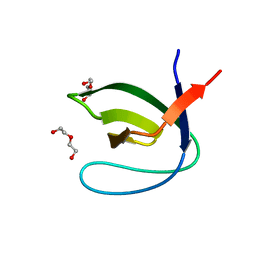 | |
7R1D
 
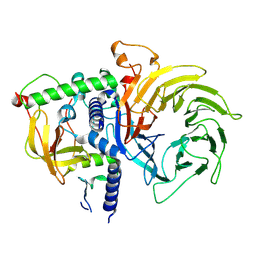 | | Structure of MuvB complex | | Descriptor: | Histone-binding protein RBBP4, Protein lin-37 homolog, Protein lin-9 homolog | | Authors: | Koliopoulos, M.G, Alfieri, C. | | Deposit date: | 2022-02-02 | | Release date: | 2022-08-10 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of a nucleosome-bound MuvB transcription factor complex reveals DNA remodelling.
Nat Commun, 13, 2022
|
|
6I3N
 
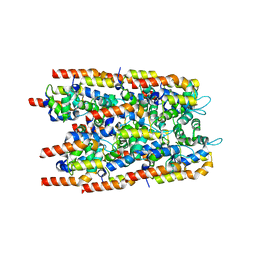 | |
6I1M
 
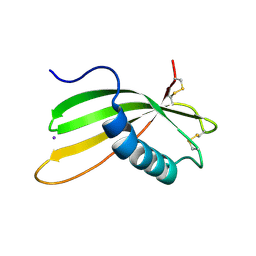 | | Secreted type 1 cystatin from Fasciola hepatica | | Descriptor: | Cystatin, IODIDE ION | | Authors: | Busa, M, Rezacova, P, Pachl, P, Stefanic, S, Mares, M. | | Deposit date: | 2018-10-29 | | Release date: | 2020-08-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An evolutionary molecular adaptation of an unusual stefin from the liver fluke Fasciola hepatica redefines the cystatin superfamily.
J.Biol.Chem., 299, 2023
|
|
7QZN
 
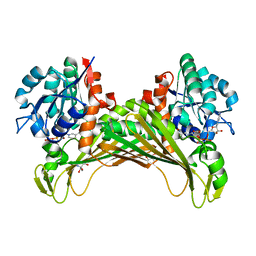 | | Amine Dehydrogenase from Cystobacter fuscus (CfusAmDH) W145A mutant with NAD+ | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Amine Dehydrogenase, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Bennett, M, Ducrot, L, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Cover Feature: Expanding the Substrate Scope of Native Amine Dehydrogenases through In Silico Structural Exploration and Targeted Protein Engineering (ChemCatChem 22/2022)
Chemcatchem, 14, 2022
|
|
7QZL
 
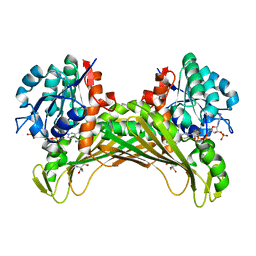 | | Amine Dehydrogenase from Cystobacter fuscus (CfusAmDH) W145A mutant with NADP+ and pentylamine | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, AMYLAMINE, Amine Dehydrogenase, ... | | Authors: | Bennett, M, Ducrot, L, Vaxelaire-Vergne, C, Grogan, G. | | Deposit date: | 2022-01-31 | | Release date: | 2022-12-07 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cover Feature: Expanding the Substrate Scope of Native Amine Dehydrogenases through In Silico Structural Exploration and Targeted Protein Engineering (ChemCatChem 22/2022)
Chemcatchem, 14, 2022
|
|
3P0H
 
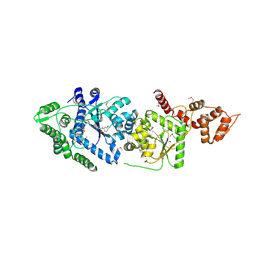 | | Leishmania major Tyrosyl-tRNA synthetase in complex with fisetin, cubic crystal form | | Descriptor: | 3,7,3',4'-TETRAHYDROXYFLAVONE, Tyrosyl-tRNA synthetase | | Authors: | Larson, E.T, Merritt, E.A, Medical Structural Genomics of Pathogenic Protozoa (MSGPP) | | Deposit date: | 2010-09-28 | | Release date: | 2011-03-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | The Double-Length Tyrosyl-tRNA Synthetase from the Eukaryote Leishmania major Forms an Intrinsically Asymmetric Pseudo-Dimer.
J.Mol.Biol., 409, 2011
|
|
1JIF
 
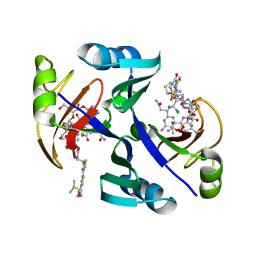 | | Crystal structure of bleomycin-binding protein from bleomycin-producing Streptomyces verticillus complexed with copper(II)-bleomycin | | Descriptor: | BLEOMYCIN A2, CHLORIDE ION, COPPER (II) ION, ... | | Authors: | Sugiyama, M, Kumagai, T, Hayashida, M, Maruyama, M, Matoba, Y. | | Deposit date: | 2001-07-02 | | Release date: | 2002-02-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The 1.6-A crystal structure of the copper(II)-bound bleomycin complexed with the bleomycin-binding protein from bleomycin-producing Streptomyces verticillus.
J.Biol.Chem., 277, 2002
|
|
4J9F
 
 | |
8UQT
 
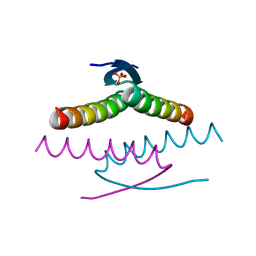 | | Crystal structure of the Tree Shrew p53 tetramerization domain | | Descriptor: | Cellular tumor antigen p53, SULFATE ION | | Authors: | Wahba, H.M, Sakaguchi, S, Nakagawa, N, Wada, J, Kamada, R, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.16 Å) | | Cite: | Highly Similar Tetramerization Domains from the p53 Protein of Different Mammalian Species Possess Varying Biophysical, Functional and Structural Properties.
Int J Mol Sci, 24, 2023
|
|
8UQS
 
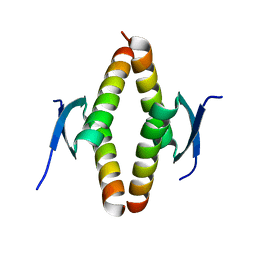 | | Crystal structure of the Opossum p53 tetramerization domain | | Descriptor: | Cellular tumor antigen p53 (Fragment) | | Authors: | Wahba, H.M, Sakaguchi, S, Nakagawa, N, Wada, J, Kamada, R, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Highly Similar Tetramerization Domains from the p53 Protein of Different Mammalian Species Possess Varying Biophysical, Functional and Structural Properties.
Int J Mol Sci, 24, 2023
|
|
8UQR
 
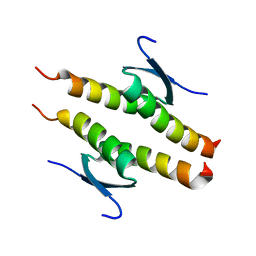 | | Crystal structure of the human p53 tetramerization domain | | Descriptor: | Cellular tumor antigen p53 | | Authors: | Wahba, H.M, Sakaguchi, S, Nakagawa, N, Wada, J, Kamada, R, Sakaguchi, K, Omichinski, J.G. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Highly Similar Tetramerization Domains from the p53 Protein of Different Mammalian Species Possess Varying Biophysical, Functional and Structural Properties.
Int J Mol Sci, 24, 2023
|
|
1BF4
 
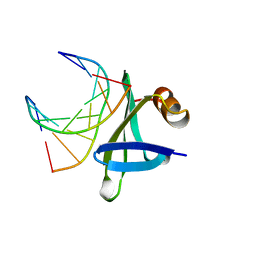 | | CHROMOSOMAL DNA-BINDING PROTEIN SSO7D/D(GCGAACGC) COMPLEX | | Descriptor: | DNA (5'-D(*GP*CP*GP*AP*AP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*TP*5IUP*CP*GP*C)-3'), PROTEIN (CHROMOSOMAL PROTEIN SSO7D) | | Authors: | Su, S, Gao, Y.-G, Robinson, H, Padmanabhan, S, Lim, L, Shriver, J.W, Wang, A.H.-J. | | Deposit date: | 1998-05-27 | | Release date: | 1999-11-10 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The crystal structure of the hyperthermophile chromosomal protein Sso7d bound to DNA.
Nat.Struct.Biol., 5, 1998
|
|
6QMJ
 
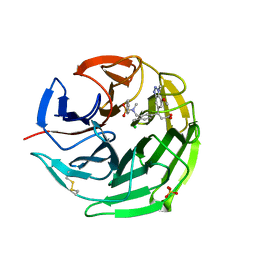 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{S})-3-(7-methoxy-1-methyl-benzotriazol-5-yl)-3-[4-methyl-3-[[methyl(phenylsulfonyl)amino]methyl]phenyl]propanoic acid, CHLORIDE ION, Kelch-like ECH-associated protein 1, ... | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6QMC
 
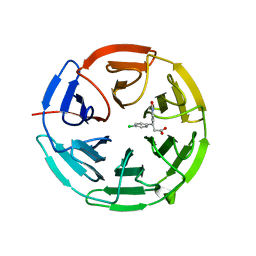 | | Small molecule inhibitor of the KEAP1-NRF2 protein-protein interaction | | Descriptor: | (3~{S})-3-(4-chlorophenyl)-3-(2-oxidanylidene-1~{H}-pyridin-4-yl)propanoic acid, Kelch-like ECH-associated protein 1 | | Authors: | Davies, T.G. | | Deposit date: | 2019-02-07 | | Release date: | 2019-04-24 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structure-Activity and Structure-Conformation Relationships of Aryl Propionic Acid Inhibitors of the Kelch-like ECH-Associated Protein 1/Nuclear Factor Erythroid 2-Related Factor 2 (KEAP1/NRF2) Protein-Protein Interaction.
J.Med.Chem., 62, 2019
|
|
6QV8
 
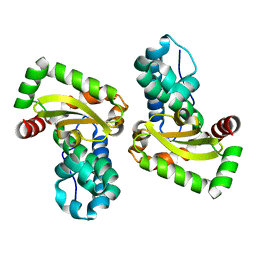 | |
6KH8
 
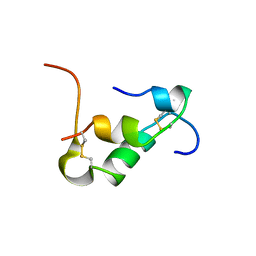 | | Solution structure of Zn free Bovine Pancreatic Insulin in 20% acetic acid-d4 (pH 1.9) | | Descriptor: | Insulin A Chain, Insulin B chain | | Authors: | Bhunia, A, Ratha, B.N, Kar, R.K, Brender, J.R. | | Deposit date: | 2019-07-14 | | Release date: | 2020-10-07 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | High-resolution structure of a partially folded insulin aggregation intermediate.
Proteins, 88, 2020
|
|
8TVZ
 
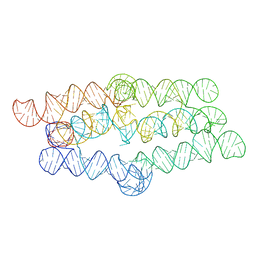 | |
2R8J
 
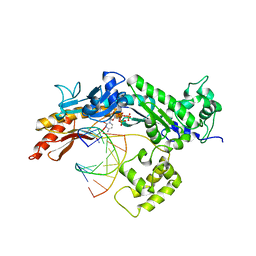 | | Structure of the Eukaryotic DNA Polymerase eta in complex with 1,2-d(GpG)-cisplatin containing DNA | | Descriptor: | 2'-DEOXYCYTIDINE-5'-TRIPHOSPHATE, CALCIUM ION, Cisplatin, ... | | Authors: | Carell, T, Alt, A, Lammens, K. | | Deposit date: | 2007-09-11 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Bypass of DNA lesions generated during anticancer treatment with cisplatin by DNA polymerase eta
Science, 318, 2007
|
|
8ABT
 
 | | Crystal structure of NaLdpA in complex with the product analog Resveratrol | | Descriptor: | RESVERATROL, SULFATE ION, SnoaL-like domain-containing protein | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6K7Y
 
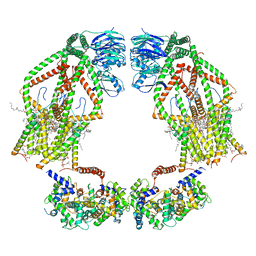 | | Intact human mitochondrial calcium uniporter complex with MICU1/MICU2 subunits | | Descriptor: | (9R,11S)-9-({[(1S)-1-HYDROXYHEXADECYL]OXY}METHYL)-2,2-DIMETHYL-5,7,10-TRIOXA-2LAMBDA~5~-AZA-6LAMBDA~5~-PHOSPHAOCTACOSANE-6,6,11-TRIOL, CALCIUM ION, CARDIOLIPIN, ... | | Authors: | Zhuo, W, Zhou, H, Yang, M. | | Deposit date: | 2019-06-10 | | Release date: | 2020-09-09 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structure of intact human MCU supercomplex with the auxiliary MICU subunits.
Protein Cell, 12, 2021
|
|
8ABV
 
 | | Crystal structure of SpLdpA in complex with erythro-DGPD | | Descriptor: | (1R,2S)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, (1S,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, GLYCEROL, ... | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.683 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8ABU
 
 | | Crystal structure of NaLdpA mutant H97Q in complex with erythro-DGPD | | Descriptor: | (1S,2R)-1,2-bis(3-methoxy-4-oxidanyl-phenyl)propane-1,3-diol, SnoaL-like domain-containing protein | | Authors: | Zahn, M, Kuatsjah, E, Beckham, G.T, McGeehan, J.E. | | Deposit date: | 2022-07-04 | | Release date: | 2023-02-01 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.661 Å) | | Cite: | Biochemical and structural characterization of a sphingomonad diarylpropane lyase for cofactorless deformylation.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
