1J3H
 
 | | Crystal structure of apoenzyme cAMP-dependent protein kinase catalytic subunit | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, cAMP-dependent protein kinase, alpha-catalytic subunit | | Authors: | Akamine, P, Madhusudan, Wu, J, Xuong, N.H, Ten Eyck, L.F, Taylor, S.S. | | Deposit date: | 2003-01-31 | | Release date: | 2003-03-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dynamic Features of cAMP-dependent Protein Kinase Revealed by Apoenzyme Crystal Structure
J.Mol.Biol., 327, 2003
|
|
1RGQ
 
 | | M9A HCV Protease complex with pentapeptide keto-amide inhibitor | | Descriptor: | N-(PYRAZIN-2-YLCARBONYL)LEUCYLISOLEUCYL-N~1~-{1-[2-({1-CARBOXY-2-[4-(PHOSPHONOOXY)PHENYL]ETHYL}AMINO)-1,1-DIHYDROXY-2-OXOETHYL]BUT-3-ENYL}-3-CYCLOHEXYLALANINAMIDE, NS3 Protease, NS4A peptide, ... | | Authors: | Liu, Y, Stoll, V.S, Richardson, P.L, Saldivar, A, Klaus, J.L, Molla, A, Kohlbrenner, W, Kati, W.M. | | Deposit date: | 2003-11-12 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Hepatitis C NS3 protease inhibition by peptidyl-alpha-ketoamide inhibitors: kinetic mechanism and structure.
Arch.Biochem.Biophys., 421, 2004
|
|
5G1B
 
 | | Bordetella Alcaligenes HDAH native | | Descriptor: | DI(HYDROXYETHYL)ETHER, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, PENTAETHYLENE GLYCOL, ... | | Authors: | Kraemer, A, Meyer-Almes, F.J, Yildiz, O. | | Deposit date: | 2016-03-24 | | Release date: | 2017-04-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The thermodynamic signature of ligand binding to histone deacetylase-like amidohydrolases is most sensitive to the flexibility in the L2-loop lining the active site pocket.
Biochim. Biophys. Acta, 1861, 2017
|
|
8VQ4
 
 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-125A. | | Descriptor: | (8R)-6-(1-benzyl-1H-pyrazole-4-carbonyl)-N-[(2S,3R)-3-(2-cyclohexylethoxy)-1-(methylamino)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Expanding the ligandable proteome by paralog hopping with covalent probes.
Biorxiv, 2024
|
|
8VQ3
 
 | | CDK2-CyclinE1 in complex with allosteric inhibitor I-198. | | Descriptor: | (8R)-N-[(2S,3R)-3-(cyclohexylmethoxy)-1-(morpholin-4-yl)-1-oxobutan-2-yl]-2-[(1S)-2,2-dimethylcyclopropane-1-carbonyl]-6-(1,3-thiazole-5-carbonyl)-2,6-diazaspiro[3.4]octane-8-carboxamide, Cyclin-dependent kinase 2, G1/S-specific cyclin-E1 | | Authors: | Hirschi, M, Johnson, E, Zhang, Y, Liu, Z, Brodsky, O, Won, S.J, Nagata, A, Petroski, M.D, Majmudar, J.D, Niessen, S, VanArsdale, T, Gilbert, A.M, Hayward, M.M, Stewart, A.E, Nager, A.R, Melillo, B, Cravatt, B. | | Deposit date: | 2024-01-17 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Expanding the ligandable proteome by paralog hopping with covalent probes.
Biorxiv, 2024
|
|
5Y38
 
 | | Crystal structure of C7orf59-HBXIP complex | | Descriptor: | Ragulator complex protein LAMTOR4, Ragulator complex protein LAMTOR5, SULFATE ION | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1
Nat Commun, 8, 2017
|
|
5Y39
 
 | | Crystal structure of Ragulator complex (p18 76-145) | | Descriptor: | Ragulator complex protein LAMTOR1, Ragulator complex protein LAMTOR2, Ragulator complex protein LAMTOR3, ... | | Authors: | Zhang, T, Ding, J. | | Deposit date: | 2017-07-28 | | Release date: | 2017-12-06 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structural basis for Ragulator functioning as a scaffold in membrane-anchoring of Rag GTPases and mTORC1
Nat Commun, 8, 2017
|
|
5GRS
 
 | | Complex structure of the fission yeast SREBP-SCAP binding domains | | Descriptor: | Sterol regulatory element-binding protein 1, Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Gong, X, Qian, H.W, Wu, J.P, Yan, N. | | Deposit date: | 2016-08-12 | | Release date: | 2016-12-14 | | Last modified: | 2019-10-23 | | Method: | ELECTRON MICROSCOPY (5.4 Å) | | Cite: | Complex structure of the fission yeast SREBP-SCAP binding domains reveals an oligomeric organization
Cell Res., 26, 2016
|
|
5GPD
 
 | |
7RWN
 
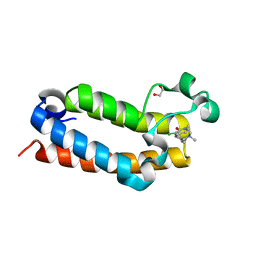 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-5-{4-[(dimethylamino)methyl]anilino}-2-methylpyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
5YDE
 
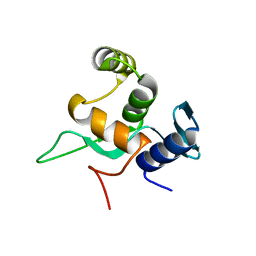 | | Crystal structure of a disease-related gene, hCDC73(1-111) | | Descriptor: | Parafibromin | | Authors: | Sun, W, Kuang, X.L, Liu, Y.P, Tian, L.F, Yan, X.X, Xu, W.Q. | | Deposit date: | 2017-09-13 | | Release date: | 2017-12-20 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.023 Å) | | Cite: | Crystal structure of the N-terminal domain of human CDC73 and its implications for the hyperparathyroidism-jaw tumor (HPT-JT) syndrome
Sci Rep, 7, 2017
|
|
7RWE
 
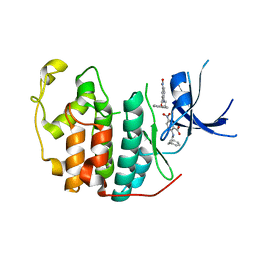 | | Crystal structure of CDK2 liganded with compound GPHR787 | | Descriptor: | 5-nitro-2-[(3-phenylpropyl)amino]benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7RWF
 
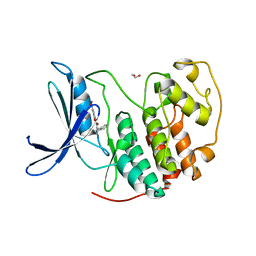 | | Crystal structure of CDK2 in complex with TW8672 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-19 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
7RWP
 
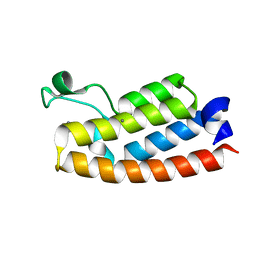 | | Crystal Structure of BPTF bromodomain in complex with 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one | | Descriptor: | 5-[4-(aminomethyl)anilino]-4-chloro-2-methylpyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWQ
 
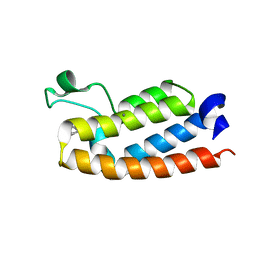 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-6-yl)amino]pyridazin-3(2H)-one, CALCIUM ION, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RWO
 
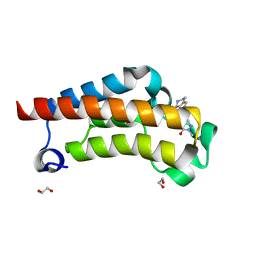 | | Crystal Structure of BPTF bromodomain in complex with 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one | | Descriptor: | 1,2-ETHANEDIOL, 4-chloro-2-methyl-5-[(1,2,3,4-tetrahydroisoquinolin-7-yl)amino]pyridazin-3(2H)-one, Nucleosome-remodeling factor subunit BPTF | | Authors: | Zahid, H, Buchholz, C, Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2021-08-20 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | New Design Rules for Developing Potent Cell-Active Inhibitors of the Nucleosome Remodeling Factor (NURF) via BPTF Bromodomain Inhibition.
J.Med.Chem., 64, 2021
|
|
7RXO
 
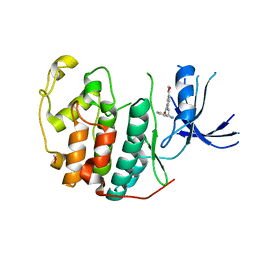 | | Crystal structure of CDK2 liganded with compound WN333 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(methoxycarbonyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-08-23 | | Release date: | 2022-08-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7S4T
 
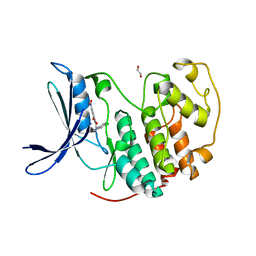 | | Crystal structure of CDK2 liganded with compound EF2252 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(6-chloro-1H-indol-3-yl)ethyl]amino}-5-nitrobenzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-09 | | Release date: | 2022-09-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7SA0
 
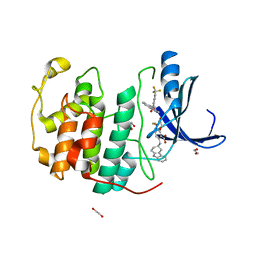 | | Crystal structure of CDK2 liganded with compound EF4195 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(3R)-2,3,4,9-tetrahydro-1H-carbazol-3-yl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-21 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Development of selective allosteric inhibitors of cyclin-dependent kinase 2 (CDK2)
To Be Published
|
|
5YBC
 
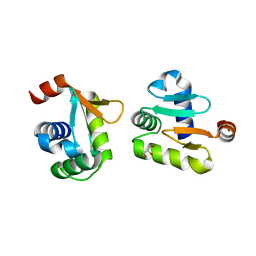 | | X-ray structure of native ETS-domain domain of Ergp55 | | Descriptor: | GLYCEROL, Transcriptional regulator ERG | | Authors: | Saxena, A.K, Gangwar, S.P. | | Deposit date: | 2017-09-04 | | Release date: | 2018-09-19 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Comparative structure analysis of the ETSi domain of ERG3 and its complex with the E74 promoter DNA sequence
Acta Crystallogr F Struct Biol Commun, 74, 2018
|
|
7S9X
 
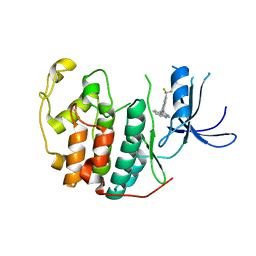 | | Crystal structure of CDK2 liganded with compound WN378 | | Descriptor: | 2-[(9H-carbazol-3-yl)amino]-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-21 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Development of selective allosteric inhibitors of cyclin-dependent kinase 2 (CDK2)
To Be Published
|
|
2M7Y
 
 | | The Mengovirus Leader protein | | Descriptor: | Leader peptide, ZINC ION | | Authors: | Cornilescu, C.C, Porter, F.W, Zhao, K.Q, Davis, V, Palmenberg, A.C, Markley, J.L. | | Deposit date: | 2013-05-02 | | Release date: | 2013-10-23 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Full Length Mengovirus Leader
To be Published
|
|
7S84
 
 | | Crystal structure of CDK2 liganded with compound TW8972 | | Descriptor: | 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Development of allosteric and selective CDK2 inhibitors for contraception with negative cooperativity to cyclin binding.
Nat Commun, 14, 2023
|
|
7S7A
 
 | | Crystal structure of CDK2 liganded with compound EF3019 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(2H-indazol-3-yl)ethyl]amino}-5-(trifluoromethyl)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-15 | | Release date: | 2022-09-28 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
7S85
 
 | | Crystal structure of CDK2 liganded with compound WN316 | | Descriptor: | 1,2-ETHANEDIOL, 2-{[2-(1H-indol-3-yl)ethyl]amino}-5-(trifluoromethoxy)benzoic acid, Cyclin-dependent kinase 2 | | Authors: | Sun, L, Schonbrunn, E. | | Deposit date: | 2021-09-17 | | Release date: | 2022-09-28 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Screening through Lead Optimization of High Affinity, Allosteric Cyclin-Dependent Kinase 2 (CDK2) Inhibitors as Male Contraceptives That Reduce Sperm Counts in Mice.
J.Med.Chem., 66, 2023
|
|
