7N3U
 
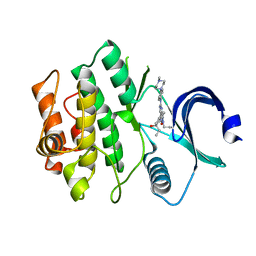 | | Crystal structure of human WEE1 kinase domain in complex with ZN-c3 | | Descriptor: | 1-[(7R)-7-ethyl-7-hydroxy-6,7-dihydro-5H-cyclopenta[b]pyridin-2-yl]-6-[4-(4-methylpiperazin-1-yl)anilino]-2-(prop-2-en-1-yl)-1,2-dihydro-3H-pyrazolo[3,4-d]pyrimidin-3-one, Wee1-like protein kinase | | Authors: | Lee, C.C. | | Deposit date: | 2021-06-02 | | Release date: | 2021-09-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Discovery of ZN-c3, a Highly Potent and Selective Wee1 Inhibitor Undergoing Evaluation in Clinical Trials for the Treatment of Cancer.
J.Med.Chem., 64, 2021
|
|
3R5N
 
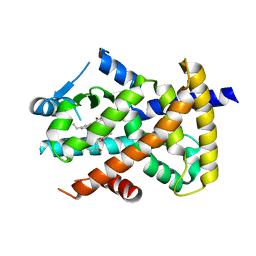 | | Crystal structure of PPARgammaLBD complexed with the agonist magnolol | | Descriptor: | 5,5'-di(prop-2-en-1-yl)biphenyl-2,2'-diol, Peroxisome proliferator-activated receptor gamma | | Authors: | Zhang, H, Chen, L, Chen, J, Hu, L, Jiang, H, Shen, X. | | Deposit date: | 2011-03-18 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular determinants of magnolol targeting both RXR(alpha) and PPAR(gamma).
Plos One, 6, 2011
|
|
5IKH
 
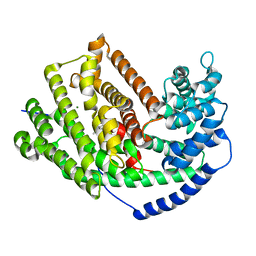 | | Tobacco 5-epi-aristolochene synthase M4 mutant with (-)-premnaspirodiene | | Descriptor: | (2R,5S,10R)-6,10-dimethyl-2-(prop-1-en-2-yl)spiro[4.5]dec-6-ene, 5-epi-aristolochene synthase, MAGNESIUM ION | | Authors: | Koo, H.J, O'Maille, P.E, Louie, G.V, Bowman, M, Noel, J.P. | | Deposit date: | 2016-03-03 | | Release date: | 2016-10-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biosynthetic potential of sesquiterpene synthases: product profiles of Egyptian Henbane premnaspirodiene synthase and related mutants.
J.Antibiot., 69, 2016
|
|
6PEL
 
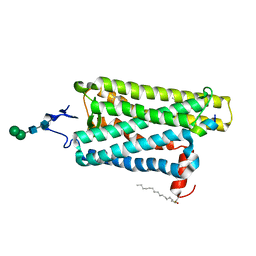 | |
6J0K
 
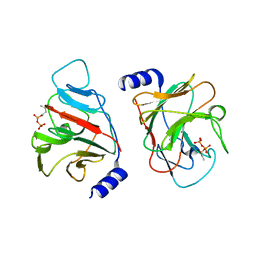 | | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A3 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
3S0E
 
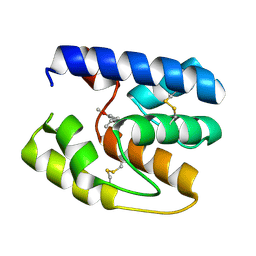 | | Apis mellifera OBP14 in complex with the odorant eugenol (2-methoxy-4(2-propenyl)-phenol) | | Descriptor: | 2-methoxy-4-(prop-2-en-1-yl)phenol, OBP14 | | Authors: | Spinelli, S, Lagarde, A, Iovinella, I, Tegoni, M, Pelosi, P, Cambillau, C. | | Deposit date: | 2011-05-13 | | Release date: | 2011-11-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Apis mellifera OBP14, a C-minus odorant-binding protein, and its complexes with odorant molecules.
Insect Biochem.Mol.Biol., 42, 2012
|
|
6P3A
 
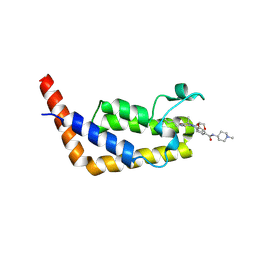 | | Crystal Structure Analysis of TAF1 Bromodomain | | Descriptor: | 4-{[(3R)-1-(but-3-en-1-yl)-3-methyl-4-(oxan-4-yl)-2-oxo-1,2,3,4-tetrahydropyrido[2,3-b]pyrazin-6-yl]amino}-3-methoxy-N-(1-methylpiperidin-4-yl)benzamide, Transcription initiation factor TFIID subunit 1 | | Authors: | Seo, H.-S, Dhe-Paganon, S. | | Deposit date: | 2019-05-23 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Crystal Structure Analysis of TAF1 Bromodomain
To Be Published
|
|
3S0K
 
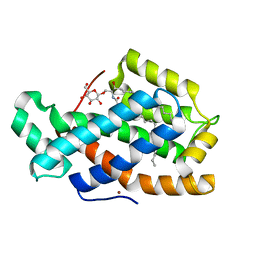 | | Crystal Structure of Human Glycolipid Transfer Protein complexed with glucosylceramide containing oleoyl acyl chain (18:1) | | Descriptor: | (9Z)-N-[(2S,3R,4E)-1-(beta-D-glucopyranosyloxy)-3-hydroxyoctadec-4-en-2-yl]octadec-9-enamide, Glycolipid transfer protein, NICKEL (II) ION | | Authors: | Cabo-Bilbao, A, Samygina, V, Popov, A.N, Ochoa-Lizarralde, B, Goni-de-Cerio, F, Patel, D.J, Brown, R.E, Malinina, L. | | Deposit date: | 2011-05-13 | | Release date: | 2012-02-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Enhanced selectivity for sulfatide by engineered human glycolipid transfer protein.
Structure, 19, 2011
|
|
6P26
 
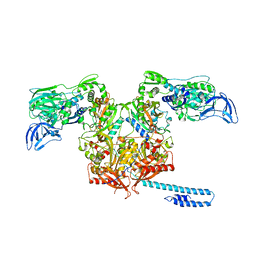 | | Escherichia coli tRNA synthetase in complex with compound 1 | | Descriptor: | N-benzyl-2-(cyclohex-1-en-1-yl)ethan-1-amine, Phenylalanine--tRNA ligase alpha subunit, Phenylalanine--tRNA ligase beta subunit | | Authors: | Kahne, D, Baidin, V, Owens, T.W. | | Deposit date: | 2019-05-21 | | Release date: | 2020-11-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.16 Å) | | Cite: | Simple Secondary Amines Inhibit Growth of Gram-Negative Bacteria through Highly Selective Binding to Phenylalanyl-tRNA Synthetase.
J.Am.Chem.Soc., 143, 2021
|
|
5YGV
 
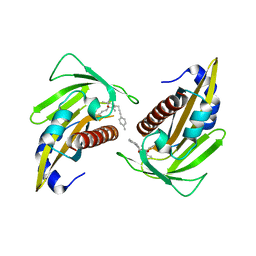 | | Crystal structure of the abscisic acid receptor PYR1 in complex with an antagonist | | Descriptor: | (2Z,4E)-3-methyl-5-[(1S,4S)-2,6,6-trimethyl-4-[3-(4-methylphenyl)prop-2-ynoxy]-1-oxidanyl-cyclohex-2-en-1-yl]penta-2,4-dienoic acid, Abscisic acid receptor PYR1 | | Authors: | Akiyama, T, Sue, M, Takeuchi, J, Mimura, N, Okamoto, M, Monda, K, Iba, K, Ohnishi, T, Todoroki, Y, Yajima, S. | | Deposit date: | 2017-09-27 | | Release date: | 2018-04-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Chemical Design of Abscisic Acid Antagonists That Block PYL-PP2C Receptor Interactions.
ACS Chem. Biol., 13, 2018
|
|
3RV7
 
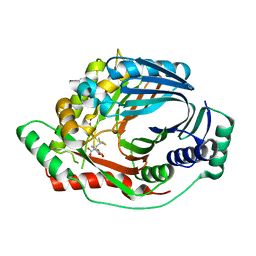 | | Structure of a M. tuberculosis Salicylate Synthase, MbtI, in Complex with an Inhibitor with Isopropyl R-Group | | Descriptor: | 3-{[(1Z)-1-carboxy-3-methylbut-1-en-1-yl]oxy}-2-hydroxybenzoic acid, Isochorismate synthase/isochorismate-pyruvate lyase mbtI | | Authors: | Chi, G, Bulloch, E.M.M, Manos-Turvey, A, Payne, R.J, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2011-05-06 | | Release date: | 2012-05-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Implications of binding mode and active site flexibility for inhibitor potency against the salicylate synthase from Mycobacterium tuberculosis
Biochemistry, 51, 2012
|
|
6J3N
 
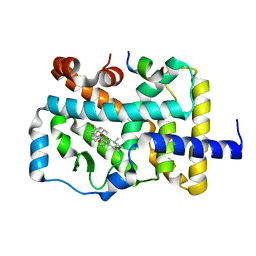 | | RORgammat LBD complexed with Ursonic Acid and SRC2.2 | | Descriptor: | (5beta)-3-oxours-12-en-28-oic acid, LYS-ILE-LEU-HIS-ARG-LEU-LEU-GLN, Nuclear receptor ROR-gamma | | Authors: | Liu, Z.H, Huang, J, Tang, Y. | | Deposit date: | 2019-01-05 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of human RORgammat LBD with SCR2.2 at 1.99 Angstroms resolution
To Be Published
|
|
7MXY
 
 | | Cryo-EM structure of PfFNT-inhibitor complex | | Descriptor: | (Z)-4,4,5,5,5-pentakis(fluoranyl)-1-(4-methoxy-2-oxidanyl-phenyl)-3-oxidanyl-pent-2-en-1-one, Formate-nitrite transporter | | Authors: | Yu, E.W, Su, C, Lyu, M. | | Deposit date: | 2021-05-19 | | Release date: | 2021-12-08 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.18 Å) | | Cite: | Structural basis of transport and inhibition of the Plasmodium falciparum transporter PfFNT
EMBO Rep, 22, 2021
|
|
7N9D
 
 | |
3S1C
 
 | | Maize cytokinin oxidase/dehydrogenase complexed with N6-isopentenyladenosine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Cytokinin dehydrogenase 1, ... | | Authors: | Kopecny, D, Briozzo, P, Morera, S. | | Deposit date: | 2011-05-15 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Kinetic and structural investigation of the cytokinin oxidase/dehydrogenase active site.
Febs J., 283, 2016
|
|
3SDC
 
 | |
3SDA
 
 | |
6H45
 
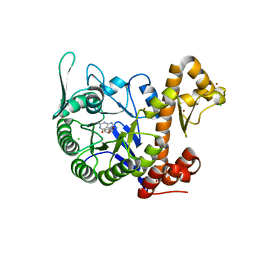 | | crystal structure of the human TGT catalytic subunit QTRT1 in complex with queuine | | Descriptor: | 2-amino-5-({[(1S,4S,5R)-4,5-dihydroxycyclopent-2-en-1-yl]amino}methyl)-3,7-dihydro-4H-pyrrolo[2,3-d]pyrimidin-4-one, BROMIDE ION, CHLORIDE ION, ... | | Authors: | Johannsson, S, Neumann, P, Ficner, R. | | Deposit date: | 2018-07-20 | | Release date: | 2018-09-05 | | Last modified: | 2023-03-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Human tRNA Guanine Transglycosylase Catalytic Subunit QTRT1.
Biomolecules, 8, 2018
|
|
5WKC
 
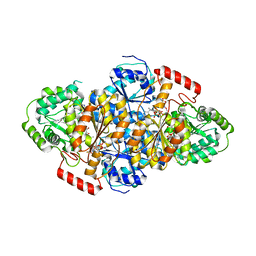 | | Saccharomyces cerevisiae acetohydroxyacid synthase in complex with the herbicide penoxsulam | | Descriptor: | (3Z)-4-{[(4-AMINO-2-METHYLPYRIMIDIN-5-YL)METHYL]AMINO}-3-MERCAPTOPENT-3-EN-1-YL TRIHYDROGEN DIPHOSPHATE, 2-(2,2-difluoroethoxy)-N-(5,8-dimethoxy[1,2,4]triazolo[1,5-c]pyrimidin-2-yl)-6-(trifluoromethyl)benzenesulfonamide, 2-[3-[(4-azanyl-2-methyl-pyrimidin-5-yl)methyl]-2-[(1~{S})-1-(dioxidanyl)-1-oxidanyl-ethyl]-4-methyl-1,3-thiazol-5-yl]ethyl phosphono hydrogen phosphate, ... | | Authors: | Guddat, W.L, Lonhienne, G.T. | | Deposit date: | 2017-07-25 | | Release date: | 2018-02-14 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | Structural insights into the mechanism of inhibition of AHAS by herbicides.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6PC6
 
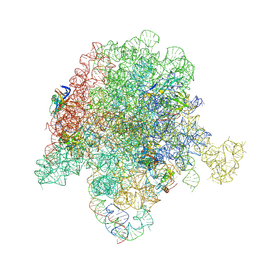 | | E. coli 50S ribosome bound to compound 47 | | Descriptor: | (3R,4R,5E,10E,12E,14S,16R,26aR)-16-fluoro-14-hydroxy-12-methyl-3-(propan-2-yl)-4-(prop-2-en-1-yl)-3,4,8,9,14,15,16,17,24,25,26,26a-dodecahydro-1H,7H,22H-21,18-(azeno)pyrrolo[2,1-c][1,8,4,19]dioxadiazacyclotetracosine-1,7,22-trione, 23S ribosomal RNA, 50S ribosomal protein L13, ... | | Authors: | Pellegrino, J, Lee, D.J, Fraser, J.S, Seiple, I.B. | | Deposit date: | 2019-06-16 | | Release date: | 2020-06-17 | | Last modified: | 2020-10-07 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Synthetic group A streptogramin antibiotics that overcome Vat resistance.
Nature, 586, 2020
|
|
5YR6
 
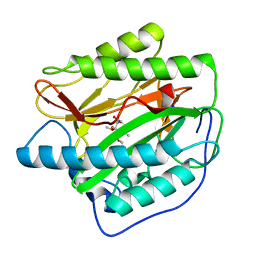 | | Human methionine aminopeptidase type 1b (F309L mutant) in complex with TNP470 | | Descriptor: | (1R,2S,3S,4R)-4-hydroxy-2-methoxy-4-methyl-3-[(2R,3R)-2-methyl-3-(3-methylbut-2-en-1-yl)oxiran-2-yl]cyclohexyl (chloroacetyl)carbamate, COBALT (II) ION, Methionine aminopeptidase 1, ... | | Authors: | Pillalamarri, V, Arya, T, Addlagatta, A. | | Deposit date: | 2017-11-08 | | Release date: | 2018-11-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Discovery of natural product ovalicin sensitive type 1 methionine aminopeptidases: molecular and structural basis.
Biochem. J., 476, 2019
|
|
3SMQ
 
 | | Crystal structure of protein arginine methyltransferase 3 | | Descriptor: | 1-(1,2,3-benzothiadiazol-6-yl)-3-[2-(cyclohex-1-en-1-yl)ethyl]urea, CHLORIDE ION, Protein arginine N-methyltransferase 3, ... | | Authors: | Dobrovetsky, E, Dong, A, Walker, J.R, Siarheyeva, A, Senisterra, G, Wasney, G.A, Smil, D, Bolshan, Y, Nguyen, K.T, Allali-Hassani, A, Hajian, T, Poda, G, Bountra, C, Weigelt, J, Edwards, A.M, Al-Awar, R, Brown, P.J, Schapira, M, Arrowsmith, C.H, Vedadi, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-06-28 | | Release date: | 2011-08-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | An allosteric inhibitor of protein arginine methyltransferase 3.
Structure, 20, 2012
|
|
6P2R
 
 | | Structure of S. cerevisiae protein O-mannosyltransferase Pmt1-Pmt2 complex bound to the sugar donor | | Descriptor: | (3R)-3,31-dimethyl-7,11,15,19,23,27-hexamethylidenedotriacont-31-en-1-yl dihydrogen phosphate, 1-PALMITOYL-2-LINOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Bai, L, Li, H. | | Deposit date: | 2019-05-21 | | Release date: | 2019-07-10 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the eukaryotic protein O-mannosyltransferase Pmt1-Pmt2 complex.
Nat.Struct.Mol.Biol., 26, 2019
|
|
6J0G
 
 | | Crystal structure of intracellular B30.2 domain of BTN3A3 mutant in complex with HMBPP | | Descriptor: | (2E)-4-hydroxy-3-methylbut-2-en-1-yl trihydrogen diphosphate, Butyrophilin subfamily 3 member A3 | | Authors: | Yang, Y.Y, Liu, W.D, Cai, N.N, Chen, C.C, Guo, R.T, Zhang, Y.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A Structural Change in Butyrophilin upon Phosphoantigen Binding Underlies Phosphoantigen-Mediated V gamma 9V delta 2 T Cell Activation.
Immunity, 50, 2019
|
|
6P5V
 
 | | Structure of DCN1 bound to N-((4S,5S)-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-1-phenyl-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl)-3-methylbenzamide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Lysozyme,DCN1-like protein 1 fusion, N-[(4S,5S)-1-[(1S)-cyclohex-3-en-1-yl]-7-ethyl-4-(4-fluorophenyl)-3-methyl-6-oxo-4,5,6,7-tetrahydro-1H-pyrazolo[3,4-b]pyridin-5-yl]-3-methylbenzamide | | Authors: | Guy, R.K, Kim, H.S, Hammill, J.T, Scott, D.C, Schulman, B.A. | | Deposit date: | 2019-05-31 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.398 Å) | | Cite: | Discovery of Novel Pyrazolo-pyridone DCN1 Inhibitors Controlling Cullin Neddylation.
J.Med.Chem., 62, 2019
|
|
