3UCX
 
 | |
3UF5
 
 | | Crystal structure of the mouse Colony-Stimulating Factor 1 (mCSF-1) cytokine | | Descriptor: | CALCIUM ION, Macrophage colony-stimulating factor 1 | | Authors: | Elegheert, J, Bracke, N, Bekaert, A, Savvides, S.N. | | Deposit date: | 2011-10-31 | | Release date: | 2012-08-22 | | Last modified: | 2013-07-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Allosteric competitive inactivation of hematopoietic CSF-1 signaling by the viral decoy receptor BARF1
Nat.Struct.Mol.Biol., 19, 2012
|
|
3SFG
 
 | |
3UVE
 
 | |
2FFY
 
 | | AmpC beta-lactamase N289A mutant in complex with a boronic acid deacylation transition state analog compound SM3 | | Descriptor: | (1R)-1-(2-THIENYLACETYLAMINO)-1-PHENYLMETHYLBORONIC ACID, Beta-lactamase, PHOSPHATE ION, ... | | Authors: | Chen, Y, Minasov, G, Roth, T.A, Prati, F, Shoichet, B.K. | | Deposit date: | 2005-12-20 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.07 Å) | | Cite: | The deacylation mechanism of AmpC beta-lactamase at ultrahigh resolution
J.Am.Chem.Soc., 128, 2006
|
|
2E1Q
 
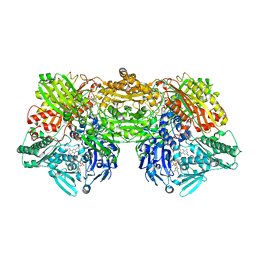 | | Crystal Structure of Human Xanthine Oxidoreductase mutant, Glu803Val | | Descriptor: | 2-HYDROXYBENZOIC ACID, BICARBONATE ION, CALCIUM ION, ... | | Authors: | Yamaguchi, Y, Matsumura, T, Ichida, K, Okamoto, K, Nishino, T. | | Deposit date: | 2006-10-27 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Human xanthine oxidase changes its substrate specificity to aldehyde oxidase type upon mutation of amino acid residues in the active site: roles of active site residues in binding and activation of purine substrate
J.Biochem.(Tokyo), 141, 2007
|
|
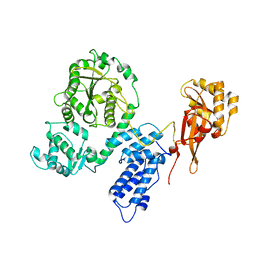
2EC5
 
 | |
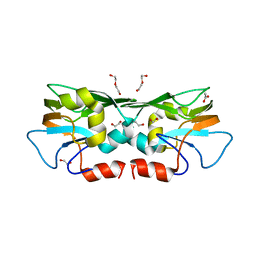
3TZU
 
 | |
3U6E
 
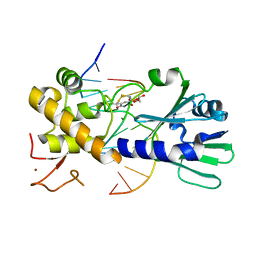 | | MutM set 1 TpGo | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*AP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*TP*(8OG)P*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3TLF
 
 | |
2F47
 
 | | Xray crystal structure of T4 lysozyme mutant L20/R63A liganded to methylguanidinium | | Descriptor: | 1-METHYLGUANIDINE, BETA-MERCAPTOETHANOL, CHLORIDE ION, ... | | Authors: | Yousef, M.S, Bischoff, N, Dyer, C.M, Baase, W.A, Matthews, B.W. | | Deposit date: | 2005-11-22 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Guanidinium derivatives bind preferentially and trigger long-distance conformational changes in an engineered T4 lysozyme.
Protein Sci., 15, 2006
|
|
3U6S
 
 | | MutM set 1 TpG | | Descriptor: | DNA (5'-D(*A*GP*GP*TP*AP*GP*AP*TP*CP*CP*AP*GP*AP*CP*GP*C)-3'), DNA (5'-D(*TP*GP*CP*GP*TP*CP*TP*GP*GP*AP*(08Q)P*CP*TP*AP*CP*C)-3'), Formamidopyrimidine-DNA glycosylase, ... | | Authors: | Sung, R.J, Zhang, M, Qi, Y, Verdine, G.L. | | Deposit date: | 2011-10-12 | | Release date: | 2012-04-25 | | Last modified: | 2013-09-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Sequence-dependent structural variation in DNA undergoing intrahelical inspection by the DNA glycosylase MutM.
J.Biol.Chem., 287, 2012
|
|
3U8B
 
 | | Functionally selective inhibition of Group IIA phospholipase A2 reveals a role for vimentin in regulating arachidonic acid metabolism | | Descriptor: | CALCIUM ION, CHLORIDE ION, Phospholipase A2, ... | | Authors: | Lee, L.K, Bryant, K.J, Bouveret, R, Lei, P.-W, Duff, A.P, Harrop, S.J, Huang, E.P, Harvey, R.P, Gelb, M.H, Gray, P.P, Curmi, P.M, Cunningham, A.M, Church, W.B, Scott, K.F. | | Deposit date: | 2011-10-16 | | Release date: | 2012-10-17 | | Last modified: | 2013-06-12 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Selective Inhibition of Human Group IIA-secreted Phospholipase A2 (hGIIA) Signaling Reveals Arachidonic Acid Metabolism Is Associated with Colocalization of hGIIA to Vimentin in Rheumatoid Synoviocytes.
J.Biol.Chem., 288, 2013
|
|
3U8J
 
 | | Crystal structure of the acetylcholine binding protein (AChBP) from Lymnaea stagnalis in complex with NS3531 (1-(pyridin-3-yl)-1,4-diazepane) | | Descriptor: | 1-(pyridin-3-yl)-1,4-diazepane, 2-acetamido-2-deoxy-beta-D-glucopyranose, Acetylcholine-binding protein, ... | | Authors: | Rohde, L.A.H, Ahring, P.K, Jensen, M.L, Nielsen, E.O, Peters, D, Helgstrand, C, Krintel, C, Harpsoe, K, Gajhede, M, Kastrup, J.S, Balle, T. | | Deposit date: | 2011-10-17 | | Release date: | 2011-12-14 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Intersubunit bridge formation governs agonist efficacy at nicotinic acetylcholine alpha 4 beta 2 receptors: unique role of halogen bonding revealed.
J.Biol.Chem., 287, 2012
|
|
3UF2
 
 | |
3V4O
 
 | |
2F2Q
 
 | | High resolution crystal structure of T4 lysozyme mutant L20R63/A liganded to guanidinium ion | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, GUANIDINE, ... | | Authors: | Yousef, M.S, Bischoff, N, Dyer, C.M, Baase, W.A, Matthews, B.W. | | Deposit date: | 2005-11-17 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Guanidinium derivatives bind preferentially and trigger long-distance conformational changes in an engineered T4 lysozyme.
Protein Sci., 15, 2006
|
|
2F32
 
 | | Xray crystal structure of lysozyme mutant L20/R63A liganded to ethylguanidinium | | Descriptor: | BETA-MERCAPTOETHANOL, Lysozyme, N-ETHYLGUANIDINE | | Authors: | Yousef, M.S, Bischoff, N, Dyer, C.M, Baase, W.A, Matthews, B.W. | | Deposit date: | 2005-11-18 | | Release date: | 2006-04-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Guanidinium derivatives bind preferentially and trigger long-distance conformational changes in an engineered T4 lysozyme.
Protein Sci., 15, 2006
|
|
2FJU
 
 | | Activated Rac1 bound to its effector phospholipase C beta 2 | | Descriptor: | 1-phosphatidylinositol-4,5-bisphosphate phosphodiesterase beta 2, 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, CALCIUM ION, ... | | Authors: | Jezyk, M.R, Snyder, J.T, Harden, T.K, Sondek, J. | | Deposit date: | 2006-01-03 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Rac1 bound to its effector phospholipase C-beta2.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2D0D
 
 | | Crystal Structure of a Meta-cleavage Product Hydrolase (CumD) A129V Mutant | | Descriptor: | 2-hydroxy-6-oxo-7-methylocta-2,4-dienoate hydrolase, CHLORIDE ION, PHOSPHATE ION | | Authors: | Jun, S.Y, Fushinobu, S, Nojiri, H, Omori, T, Shoun, H, Wakagi, T. | | Deposit date: | 2005-08-01 | | Release date: | 2006-06-06 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Improving the catalytic efficiency of a meta-cleavage product hydrolase (CumD) from Pseudomonas fluorescens IP01
Biochim.Biophys.Acta, 1764, 2006
|
|
3V4L
 
 | |
3V55
 
 | | Human MALT1 (334-719) in its ligand free form | | Descriptor: | Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | Renatus, M, Wiesmann, C. | | Deposit date: | 2011-12-16 | | Release date: | 2012-03-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Determinants of MALT1 Protease Activity.
J.Mol.Biol., 419, 2012
|
|
2CW2
 
 | | Crystal structure of Superoxide dismutase from P. Marinus | | Descriptor: | FE (III) ION, superoxide dismutase 1 | | Authors: | Asojo, O.A, Schott, E.J, Vasta, G.R, Silva, A.M. | | Deposit date: | 2005-06-16 | | Release date: | 2006-07-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structures of PmSOD1 and PmSOD2, two superoxide dismutases from the protozoan parasite Perkinsus marinus
ACTA CRYSTALLOGR.,SECT.F, 62, 2006
|
|
2FPB
 
 | |
2FP8
 
 | |
