3X1M
 
 | |
3X1K
 
 | |
1MAB
 
 | | RAT LIVER F1-ATPASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bianchet, M.A, Amzel, L.M. | | Deposit date: | 1998-08-06 | | Release date: | 1998-09-30 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The 2.8-A structure of rat liver F1-ATPase: configuration of a critical intermediate in ATP synthesis/hydrolysis.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
1EFC
 
 | | INTACT ELONGATION FACTOR FROM E.COLI | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, PROTEIN (ELONGATION FACTOR) | | Authors: | Song, H, Parsons, M.R, Rowsell, S, Leonard, G, Phillips, S.E.V. | | Deposit date: | 1998-11-24 | | Release date: | 1999-03-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of intact elongation factor EF-Tu from Escherichia coli in GDP conformation at 2.05 A resolution.
J.Mol.Biol., 285, 1999
|
|
1VA6
 
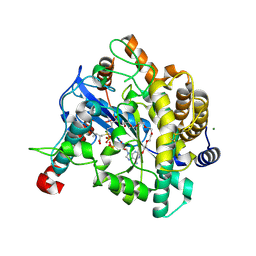 | | Crystal structure of Gamma-glutamylcysteine synthetase from Escherichia Coli B complexed with Transition-state analogue | | Descriptor: | (2S)-2-AMINO-4-[[(2R)-2-CARBOXYBUTYL](PHOSPHONO)SULFONIMIDOYL]BUTANOIC ACID, ADENOSINE-5'-DIPHOSPHATE, Glutamate--cysteine ligase, ... | | Authors: | Hibi, T, Nii, H, Nakatsu, T, Kato, H, Hiratake, J, Oda, J. | | Deposit date: | 2004-02-12 | | Release date: | 2004-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of gamma-glutamylcysteine synthetase: insights into the mechanism of catalysis by a key enzyme for glutathione homeostasis
PROC.NATL.ACAD.SCI.USA, 101, 2004
|
|
4XR8
 
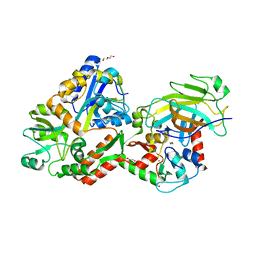 | | Crystal structure of the HPV16 E6/E6AP/p53 ternary complex at 2.25 A resolution | | Descriptor: | 1,2-ETHANEDIOL, Cellular tumor antigen p53, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Martinez-Zapien, D, Ruiz, F.X, Mitschler, A, Podjarny, A, Trave, G, Zanier, K. | | Deposit date: | 2015-01-20 | | Release date: | 2016-02-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure of the E6/E6AP/p53 complex required for HPV-mediated degradation of p53.
Nature, 529, 2016
|
|
1EPU
 
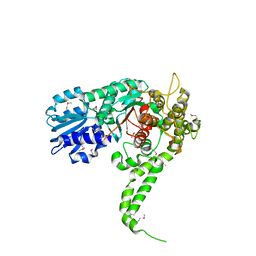 | | X-RAY crystal structure of neuronal SEC1 from squid | | Descriptor: | S-SEC1 | | Authors: | Bracher, A, Perrakis, A, Dresbach, T, Betz, H, Weissenhorn, W. | | Deposit date: | 2000-03-29 | | Release date: | 2000-08-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The X-ray crystal structure of neuronal Sec1 from squid sheds new light on the role of this protein in exocytosis.
Structure Fold.Des., 8, 2000
|
|
8DSO
 
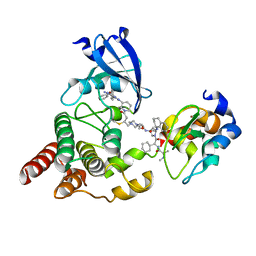 | | Structure of cIAP1, BTK and BCCov | | Descriptor: | (4S)-4-[2-(2-{4-[(2E)-4-{(3R)-3-[4-amino-3-(4-phenoxyphenyl)-1H-pyrazolo[3,4-d]pyrimidin-1-yl]piperidin-1-yl}-4-oxobut-2-en-1-yl]piperazin-1-yl}ethoxy)acetamido]-1-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]-L-prolinamide bound form, Baculoviral IAP repeat-containing protein 2, Tyrosine-protein kinase BTK, ... | | Authors: | Schiemer, J.S, Calabrese, M.F. | | Deposit date: | 2022-07-22 | | Release date: | 2023-03-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.334 Å) | | Cite: | A covalent BTK ternary complex compatible with targeted protein degradation.
Nat Commun, 14, 2023
|
|
2DFY
 
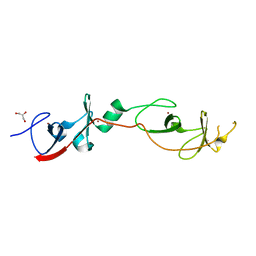 | | Crystal structure of a cyclized protein fusion of LMO4 LIM domains 1 and 2 with the LIM interacting domain of LDB1 | | Descriptor: | GLYCEROL, ZINC ION, fusion protein of LIM domain transcription factor LMO4 and LIM domain-binding protein 1 | | Authors: | Jeffries, C.M.J, Graham, S.C, Collyer, C.A, Guss, J.M, Matthews, J.M. | | Deposit date: | 2006-03-06 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Stabilization of a binary protein complex by intein-mediated cyclization
Protein Sci., 15, 2006
|
|
3P6J
 
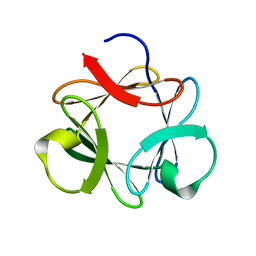 | |
3Q7Y
 
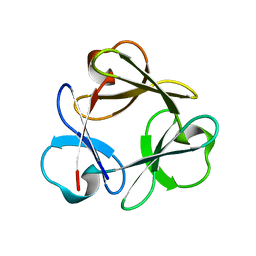 | |
3Q7W
 
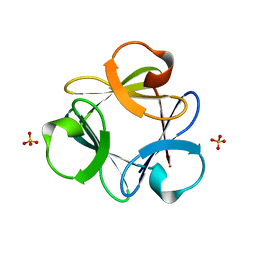 | |
5NKL
 
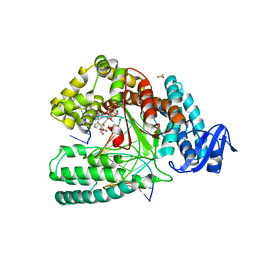 | | Crystal structure of the large fragment of DNA polymerase I from Thermus Aquaticus in a closed ternary complex with the artificial base pair dDs-dPxTP | | Descriptor: | ACETATE ION, DNA (5'-D(*AP*AP*AP*(DNU)P*GP*GP*CP*GP*CP*CP*GP*TP*GP*GP*TP*C)-3'), DNA (5'-D(*GP*AP*CP*CP*AP*CP*GP*GP*CP*GP*CP*(DOC))-3'), ... | | Authors: | Betz, K, Marx, A, Diederichs, K, Hirao, I, Kimoto, M. | | Deposit date: | 2017-03-31 | | Release date: | 2017-06-28 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural Basis for Expansion of the Genetic Alphabet with an Artificial Nucleobase Pair.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
4IOB
 
 | |
8PZG
 
 | |
6Y8J
 
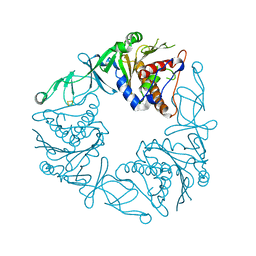 | | Crystal structure of the apo form of a quaternary ammonium Rieske monooxygenase CntA | | Descriptor: | Carnitine monooxygenase oxygenase subunit, FE2/S2 (INORGANIC) CLUSTER | | Authors: | Quareshy, M, Shanmugam, M, Bugg, T.D, Cameron, A, Chen, Y. | | Deposit date: | 2020-03-05 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis of carnitine monooxygenase CntA substrate specificity, inhibition, and intersubunit electron transfer.
J.Biol.Chem., 296, 2020
|
|
4XSS
 
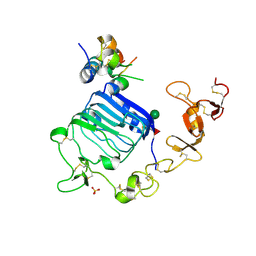 | | Insulin-like growth factor I in complex with site 1 of a hybrid insulin receptor / Type 1 insulin-like growth factor receptor | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Insulin receptor, ... | | Authors: | Lawrence, C, Kong, G.K.-W, Menting, J.G, Lawrence, M.C. | | Deposit date: | 2015-01-22 | | Release date: | 2015-06-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural Congruency of Ligand Binding to the Insulin and Insulin/Type 1 Insulin-like Growth Factor Hybrid Receptors.
Structure, 23, 2015
|
|
1H8A
 
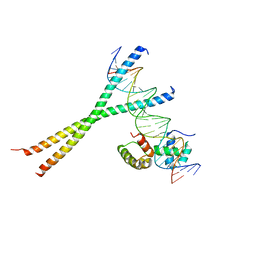 | | CRYSTAL STRUCTURE OF TERNARY PROTEIN-DNA COMPLEX3 | | Descriptor: | CAAT/ENHANCER BINDING PROTEIN BETA, DNA(5'-(*CP*CP*AP*GP*TP*CP*CP*GP*TP*TP*AP* AP*GP*GP*AP*TP*TP*GP*CP*GP*CP*CP*AP*CP*AP*T)-3'), DNA(5'-(*GP*AP*TP*GP*TP*GP*GP*CP*GP*CP*AP* AP*TP*CP*CP*TP*TP*AP*AP*CP*GP*GP*AP*CP*TP*G)-3'), ... | | Authors: | Tahirov, T.H, Ogata, K. | | Deposit date: | 2001-01-31 | | Release date: | 2002-01-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.23 Å) | | Cite: | Mechanism of C-Myb-C/Ebpbeta Cooperation from Separated Sites on a Promoter
Cell(Cambridge,Mass.), 108, 2002
|
|
1ABR
 
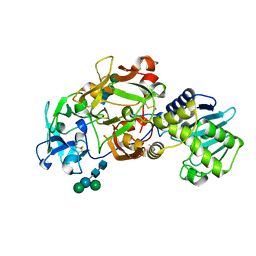 | | CRYSTAL STRUCTURE OF ABRIN-A | | Descriptor: | ABRIN-A, beta-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-alpha-L-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, beta-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Tahirov, T.H, Lu, T.-H, Liaw, Y.-C, Chu, S.-C, Lin, J.-Y. | | Deposit date: | 1994-11-11 | | Release date: | 1995-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Crystal structure of abrin-a at 2.14 A.
J.Mol.Biol., 250, 1995
|
|
5OOG
 
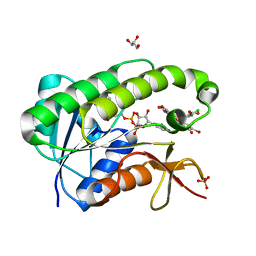 | | Human biliverdin IX beta reductase: NADP/Phloxine B ternary complex | | Descriptor: | Flavin reductase (NADPH), GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Manso, J.A, Pereira, P.J.B. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | In silicoand crystallographic studies identify key structural features of biliverdin IX beta reductase inhibitors having nanomolar potency.
J. Biol. Chem., 293, 2018
|
|
2FDE
 
 | | Wild type HIV protease bound with GW0385 | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL [(1S,2R)-3-[(1,3-BENZODIOXOL-5-YLSULFONYL)(ISOBUTYL)AMINO]-2-HYDROXY-1-{4-[(2-METHYL-1,3-THIAZOL-4-YL)METHOXY]BENZYL}PROPYL]CARBAMATE, Gag-Pol polyprotein, POTASSIUM ION | | Authors: | Xu, R.X. | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ultra-potent P1 modified arylsulfonamide HIV protease inhibitors: The discovery of GW0385.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
4AP0
 
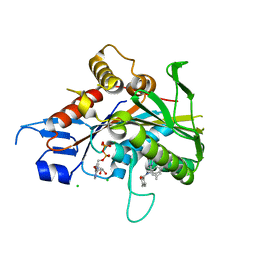 | | The mitotic kinesin Eg5 in complex with Mg-ADP and ispinesib | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, ISPINESIB MESILATE, ... | | Authors: | Schuettelkopf, A.W, Talapatra, S.K, Kozielski, F. | | Deposit date: | 2012-03-30 | | Release date: | 2012-09-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.594 Å) | | Cite: | The Structure of the Ternary Eg5-Adp-Ispinesib Complex
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4Y5X
 
 | | Diabody 305 complex with EpoR | | Descriptor: | CITRATE ANION, DI(HYDROXYETHYL)ETHER, Erythropoietin receptor, ... | | Authors: | Moraga, I, Guo, F, Ozkan, E, Jude, K.M, Garcia, K.C. | | Deposit date: | 2015-02-12 | | Release date: | 2015-03-18 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Tuning Cytokine Receptor Signaling by Re-orienting Dimer Geometry with Surrogate Ligands.
Cell, 160, 2015
|
|
8ETD
 
 | | Crystal Structure of Schizosaccharomyces pombe Rho1 | | Descriptor: | GTP-binding protein rho1, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION | | Authors: | Huang, Q, Xie, J, Seetharaman, J. | | Deposit date: | 2022-10-16 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Crystal Structure of Schizosaccharomyces pombe Rho1 Reveals Its Evolutionary Relationship with Other Rho GTPases.
Biology (Basel), 11, 2022
|
|
3Q7X
 
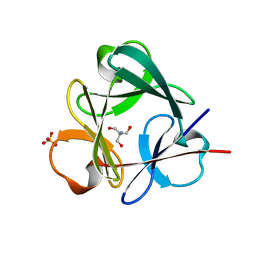 | | Crystal structure of Symfoil-4P/PV1: de novo designed beta-trefoil architecture with symmetric primary structure, primitive version 1 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, SULFATE ION, de novo designed beta-trefoil architecture with symmetric primary structure | | Authors: | Blaber, M, Lee, J. | | Deposit date: | 2011-01-05 | | Release date: | 2012-01-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Simplified protein design biased for prebiotic amino acids yields a foldable, halophilic protein.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
