3I4V
 
 | | Crystal structure determination of catechol 1,2-dioxygenase from rhodococcus opacus 1CP in complex with 3-chlorocatechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 3-chlorobenzene-1,2-diol, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-07-03 | | Release date: | 2010-01-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
3I51
 
 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with 4,5-dichlorocatechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 4,5-dichlorobenzene-1,2-diol, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Kolomytseva, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-07-03 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
3I5H
 
 | | The crystal structure of rigor like squid myosin S1 in the absence of nucleotide | | Descriptor: | CALCIUM ION, Myosin catalytic light chain LC-1, mantle muscle, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
7JGM
 
 | | Crystal Structure of the Ni-bound Human Heavy-chain variant 122H-delta C-star with meta-benzenedihyrdoxamate | | Descriptor: | Ferritin heavy chain, NICKEL (II) ION, N~1~,N~3~-dihydroxybenzene-1,3-dicarboxamide, ... | | Authors: | Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2020-07-19 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Tunable and Cooperative Thermomechanical Properties of Protein-Metal-Organic Frameworks.
J.Am.Chem.Soc., 142, 2020
|
|
4ZBB
 
 | |
3I5F
 
 | | Crystal structure of squid MG.ADP myosin S1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Myosin catalytic light chain LC-1, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-08-04 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
6RFH
 
 | |
3I5G
 
 | | Crystal structure of rigor-like squid myosin S1 | | Descriptor: | CALCIUM ION, MALONATE ION, Myosin catalytic light chain LC-1, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
3I4Y
 
 | | Crystal structure determination of Catechol 1,2-Dioxygenase from Rhodococcus opacus 1CP in complex with 3,5-dichlorocatechol | | Descriptor: | (4S,7R)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSAN-1-AMINIUM 4-OXIDE, 3,5-dichlorobenzene-1,2-diol, Catechol 1,2-dioxygenase, ... | | Authors: | Matera, I, Ferraroni, M, Kolomytseva, M, Briganti, F, Scozzafava, A. | | Deposit date: | 2009-07-03 | | Release date: | 2010-01-12 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Catechol 1,2-dioxygenase from the Gram-positive Rhodococcus opacus 1CP: Quantitative structure/activity relationship and the crystal structures of native enzyme and catechols adducts.
J.Struct.Biol., 170, 2010
|
|
6UIK
 
 | |
3I5I
 
 | | The crystal structure of squid myosin S1 in the presence of SO4 2- | | Descriptor: | CALCIUM ION, Myosin catalytic light chain LC-1, mantle muscle, ... | | Authors: | Yang, Y, Gourinath, S, Kovacs, M, Nyitray, L, Reutzel, R, Himmel, D.M, O'Neall-Hennessey, E, Reshetnikova, L, Szent-Gyorgyi, A.G, Brown, J.H, Cohen, C. | | Deposit date: | 2009-07-05 | | Release date: | 2009-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Rigor-like structures from muscle myosins reveal key mechanical elements in the transduction pathways of this allosteric motor.
Structure, 15, 2007
|
|
6UE5
 
 | | Crystal structure of full-length human DCAF15-DDB1-deltaPBP-DDA1-RBM39 in complex with 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzenesulfonamide | | Descriptor: | 4-(aminomethyl)-N-(3-cyano-4-methyl-1H-indol-7-yl)benzene-1-sulfonamide, DDB1- and CUL4-associated factor 15, DET1- and DDB1-associated protein 1, ... | | Authors: | Knapp, M.S, Shu, W, Xie, L, Bussiere, D.E. | | Deposit date: | 2019-09-20 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Structural basis of indisulam-mediated RBM39 recruitment to DCAF15 E3 ligase complex.
Nat.Chem.Biol., 16, 2020
|
|
6UJ4
 
 | |
6E69
 
 | | Ortho-substituted phenyl sulfonyl fluoride and fluorosulfate as potent elastase inhibitory fragments | | Descriptor: | 2-(fluorosulfonyl)benzene-1-sulfonic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wolan, D.W, Woehl, J.L, Kitamura, S. | | Deposit date: | 2018-07-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | SuFEx-enabled, agnostic discovery of covalent inhibitors of human neutrophil elastase.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
8FO3
 
 | |
6SBH
 
 | | Human Carbonic Anhydrase II in complex with 4-pentylbenzenesulfonamide. | | Descriptor: | (4-CARBOXYPHENYL)(CHLORO)MERCURY, 4-pentylbenzenesulfonamide, Carbonic anhydrase 2, ... | | Authors: | Gloeckner, S, Ngo, K, Heine, A, Klebe, G. | | Deposit date: | 2019-07-20 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.95 Å) | | Cite: | Conformational Changes in Alkyl Chains Determine the Thermodynamic and Kinetic Binding Profiles of Carbonic Anhydrase Inhibitors.
Acs Chem.Biol., 15, 2020
|
|
7JGN
 
 | | Crystal Structure of the Zn-bound Human Heavy-chain variant 122H-delta C-star with meta-benzenedihyrdoxamate collected at 100K | | Descriptor: | Ferritin heavy chain, N~1~,N~3~-dihydroxybenzene-1,3-dicarboxamide, SODIUM ION, ... | | Authors: | Bailey, J.B, Tezcan, F.A. | | Deposit date: | 2020-07-19 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Tunable and Cooperative Thermomechanical Properties of Protein-Metal-Organic Frameworks.
J.Am.Chem.Soc., 142, 2020
|
|
6UOZ
 
 | |
7MYD
 
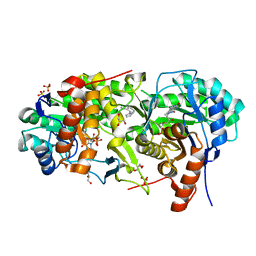 | | Leishmania major dihydroorotate dehydrogenase in complex with 5-amino-2-(1H-pyrrol-1-yl)benzonitrile | | Descriptor: | 5-azanyl-2-pyrrol-1-yl-benzenecarbonitrile, Dihydroorotate dehydrogenase (fumarate), FLAVIN MONONUCLEOTIDE, ... | | Authors: | Pinheiro, M.P, Cardoso, I.A, Hunter, W.N, Nonato, M.C. | | Deposit date: | 2021-05-20 | | Release date: | 2022-05-25 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Leishmania major dihydroorotate dehydrogenase in complex with 5-amino-2-(1H-pyrrol-1-yl)benzonitrile
To Be Published
|
|
7AES
 
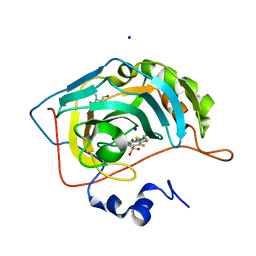 | | Human carbonic anhydrase II in complex with 2,3,5,6-tetrafluoro-N-methyl-4-propylsulfanyl-benzenesulfonamide | | Descriptor: | 2,3,5,6-tetrakis(fluoranyl)-~{N}-methyl-4-propylsulfanyl-benzenesulfonamide, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, BICINE, ... | | Authors: | Paketuryte, V, Smirnov, A, Manakova, E, Grazulis, S. | | Deposit date: | 2020-09-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and mechanism of secondary sulfonamide binding to carbonic anhydrases.
Eur.Biophys.J., 50, 2021
|
|
1VYZ
 
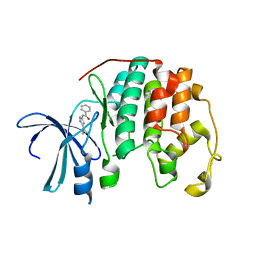 | | Structure of CDK2 complexed with PNU-181227 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, N-(5-CYCLOPROPYL-1H-PYRAZOL-3-YL)BENZAMIDE | | Authors: | Pevarello, P, Brasca, M.G, Amici, R, Orsini, P, Traquandi, G, Corti, L, Piutti, C, Sansonna, P, Villa, M, Pierce, B.S, Pulici, M, Giordano, G, Martina, K, Lfritzen, E, Nugent, R.A, Casale, E, Cameron, A, Ciomei, M, Roletto, F, Isacchi, A, Fogliatto, G, Pesenti, E, Pastori, W, Marsiglio, W, Leach, K.L, Clare, P.M, Fiorentini, F, Varasi, M, Vulpetti, A, Warpehoski, M.A. | | Deposit date: | 2004-05-07 | | Release date: | 2004-06-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | 3-Aminopyrazole Inhibitors of Cdk2/Cyclin a as Antitumor Agents. 1. Lead Finding
J.Med.Chem., 47, 2004
|
|
5J27
 
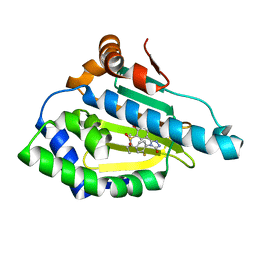 | | HSP90 in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-methyl-N-propyl-benzenesulfonamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methyl-N-propylbenzene-1-sulfonamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-03-29 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
1S5G
 
 | | Structure of Scallop myosin S1 reveals a novel nucleotide conformation | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Risal, D, Gourinath, S, Himmel, D.M, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2004-01-20 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1SR6
 
 | | Structure of nucleotide-free scallop myosin S1 | | Descriptor: | CALCIUM ION, MAGNESIUM ION, Myosin essential light chain, ... | | Authors: | Risal, D, Gourinath, S, Himmel, D.M, Szent-Gyorgyi, A.G, Cohen, C. | | Deposit date: | 2004-03-22 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Myosin subfragment 1 structures reveal a partially bound nucleotide and a complex salt bridge that helps couple nucleotide and actin binding.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
5J6N
 
 | | Crystal Structure of Hsp90-alpha N-domain L107A mutant in complex with 5-[4-(2-Fluoro-phenyl)-5-oxo-4,5-dihydro-1H-[1,2,4]triazol-3-yl]-2,4-dihydroxy-N-methyl-N-propyl-benzenesulfonamide | | Descriptor: | 5-[4-(2-fluorophenyl)-5-oxo-4,5-dihydro-1H-1,2,4-triazol-3-yl]-2,4-dihydroxy-N-methyl-N-propylbenzene-1-sulfonamide, Heat shock protein HSP 90-alpha | | Authors: | Amaral, M, Matias, P. | | Deposit date: | 2016-04-05 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Protein conformational flexibility modulates kinetics and thermodynamics of drug binding.
Nat Commun, 8, 2017
|
|
