6GYU
 
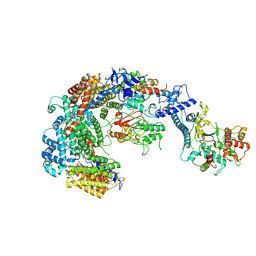 | | Cryo-EM structure of the CBF3-msk complex of the budding yeast kinetochore | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-02 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GYS
 
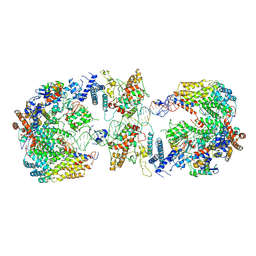 | | Cryo-EM structure of the CBF3-CEN3 complex of the budding yeast kinetochore | | Descriptor: | Centromere DNA-binding protein complex CBF3 subunit A, Centromere DNA-binding protein complex CBF3 subunit B, Centromere DNA-binding protein complex CBF3 subunit C, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-01 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6GYP
 
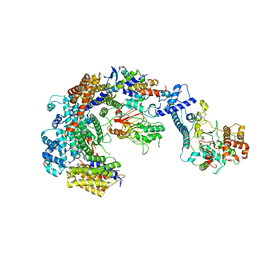 | | Cryo-EM structure of the CBF3-core-Ndc10-DBD complex of the budding yeast kinetochore | | Descriptor: | ARGININE, ASPARAGINE, Centromere DNA-binding protein complex CBF3 subunit A, ... | | Authors: | Yan, K, Zhang, Z, Yang, J, McLaughlin, S.H, Barford, D. | | Deposit date: | 2018-07-01 | | Release date: | 2018-12-05 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Architecture of the CBF3-centromere complex of the budding yeast kinetochore.
Nat. Struct. Mol. Biol., 25, 2018
|
|
6HCY
 
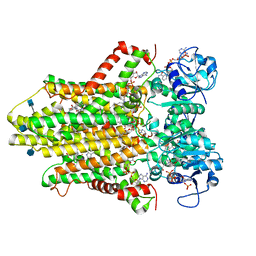 | | human STEAP4 bound to NADP, FAD, heme and Fe(III)-NTA. | | Descriptor: | (2R)-3-(phosphonooxy)propane-1,2-diyl dihexanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Oosterheert, W, van Bezouwen, L.S, Rodenburg, R.N.P, Forster, F, Mattevi, A, Gros, P. | | Deposit date: | 2018-08-17 | | Release date: | 2018-10-24 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Cryo-EM structures of human STEAP4 reveal mechanism of iron(III) reduction.
Nat Commun, 9, 2018
|
|
9H4R
 
 | |
9H4S
 
 | |
4V6A
 
 | | Structure of EF-P bound to the 70S ribosome. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Stanley, R.E, Blaha, G. | | Deposit date: | 2009-06-15 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Formation of the first peptide bond: the structure of EF-P bound to the 70S ribosome.
Science, 325, 2009
|
|
4V7O
 
 | | Proteasome Activator Complex | | Descriptor: | Proteasome activator BLM10, Proteasome component C1, Proteasome component C11, ... | | Authors: | Hill, C.P, Whitby, F.G. | | Deposit date: | 2009-12-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening.
Mol.Cell, 37, 2010
|
|
6OVH
 
 | | Cryo-EM structure of Bimetallic dodecameric cage design 3 (BMC3) from cytochrome cb562 | | Descriptor: | ACETOHYDROXAMIC ACID, FE (III) ION, HEME C, ... | | Authors: | Golub, E, Subramanian, R.H, Yan, X, Alberstein, R.G, Tezcan, F.A. | | Deposit date: | 2019-05-07 | | Release date: | 2020-01-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Constructing protein polyhedra via orthogonal chemical interactions.
Nature, 578, 2020
|
|
4V9A
 
 | | Crystal Structure of the 70S ribosome with tetracycline. | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Jenner, L, Yusupov, M, Yusupova, G. | | Deposit date: | 2012-07-18 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2999 Å) | | Cite: | Structural basis for potent inhibitory activity of the antibiotic tigecycline during protein synthesis.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4W2E
 
 | | Crystal structure of Elongation Factor 4 (EF4/LepA) bound to the Thermus thermophilus 70S ribosome | | Descriptor: | 16S Ribosomal RNA, 23S Ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Gagnon, M.G, Lin, J, Steitz, T.A. | | Deposit date: | 2014-06-04 | | Release date: | 2014-10-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of elongation factor 4 bound to a clockwise ratcheted ribosome.
Science, 345, 2014
|
|
8CNZ
 
 | | mmLarE-[4Fe-4S] phased by Fe-SAD | | Descriptor: | CHLORIDE ION, IODIDE ION, IRON/SULFUR CLUSTER, ... | | Authors: | Pecqueur, L, Zecchin, P, Golinelli-Pimpaneau, B. | | Deposit date: | 2023-02-24 | | Release date: | 2024-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structure-based insights into the mechanism of [4Fe-4S]-dependent sulfur insertase LarE.
Protein Sci., 33, 2024
|
|
6P18
 
 | | Q21 transcription antitermination complex: loading complex | | Descriptor: | DNA (67-MER) fragment carrying phage-21 pR' promoter and pause element, nontemplate strand, template strand, ... | | Authors: | Yin, Z, Ebright, R.H. | | Deposit date: | 2019-05-19 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of Q-dependent antitermination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4W9Q
 
 | | The Fk1 domain of FKBP51 in complex with (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-5-ethyl-3,10-diazabicyclo[4.3.1]decan-2-one | | Descriptor: | (1S,5S,6R)-10-[(3,5-dichlorophenyl)sulfonyl]-3-[2-(3,4-dimethoxyphenoxy)ethyl]-5-ethyl-3,10-diazabicyclo[4.3.1]decan-2-one, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Pomplun, S, Wang, Y, Kirschner, K, Kozany, C, Bracher, A, Hausch, F. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | Rational Design and Asymmetric Synthesis of Potent and Neurotrophic Ligands for FK506-Binding Proteins (FKBPs).
Angew.Chem.Int.Ed.Engl., 54, 2015
|
|
4V8F
 
 | | Crystal structure analysis of ribosomal decoding (near-cognate tRNA-ttyr complex with paromomycin). | | Descriptor: | 16S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Jenner, L, Demeshkina, N, Yusupov, M, Yusupova, G. | | Deposit date: | 2011-12-07 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A new understanding of the decoding principle on the ribosome.
Nature, 484, 2012
|
|
4WF7
 
 | | Crystal structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in the intramolecular isomerization catalysis | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wang, Y.L, Chow, S.Y, Lin, Y.T, Hsieh, Y.C, Lee, G.C, Liaw, S.H. | | Deposit date: | 2014-09-13 | | Release date: | 2014-12-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Structures of trehalose synthase from Deinococcus radiodurans reveal that a closed conformation is involved in catalysis of the intramolecular isomerization.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4WPZ
 
 | | Crystal structure of cytochrome P450 CYP107W1 from Streptomyces avermitilis | | Descriptor: | Cytochrome P450, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kang, L.W, Kim, D.H, Pham, T.V, Han, S.H. | | Deposit date: | 2014-10-21 | | Release date: | 2015-04-22 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Functional characterization of CYP107W1 from Streptomyces avermitilis and biosynthesis of macrolide oligomycin A.
Arch.Biochem.Biophys., 575, 2015
|
|
5V10
 
 | | Crystal structure of the putative tol-pal system-associated acyl-CoA thioesterase from Pseudomonas aeruginosa PAO1 | | Descriptor: | CHLORIDE ION, Uncharacterized protein | | Authors: | Borek, D, Wawrzak, Z, Grimshaw, S, Sandoval, J, Evdokimova, E, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-28 | | Release date: | 2017-03-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the putative tol-pal system-associated acyl-CoA thioesterase from Pseudomonas aeruginosa PAO1
To Be Published
|
|
7SKS
 
 | |
7SKU
 
 | | Nipah virus matrix protein in complex with PI(4,5)P2 | | Descriptor: | Matrix protein, SULFATE ION, [(2R)-2-octanoyloxy-3-[oxidanyl-[(1R,2R,3S,4R,5R,6S)-2,3,6-tris(oxidanyl)-4,5-diphosphonooxy-cyclohexyl]oxy-phosphoryl]oxy-propyl] octanoate | | Authors: | Norris, M.J, Saphire, E.O. | | Deposit date: | 2021-10-21 | | Release date: | 2022-08-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.117 Å) | | Cite: | Measles and Nipah virus assembly: Specific lipid binding drives matrix polymerization.
Sci Adv, 8, 2022
|
|
7SKT
 
 | |
6PPB
 
 | | Kaposi's sarcoma-associated herpesvirus (KSHV), C5 portal vertex structure | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Gong, D, Dai, X, Jih, J, Liu, Y.T, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2019-07-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | DNA-Packing Portal and Capsid-Associated Tegument Complexes in the Tumor Herpesvirus KSHV.
Cell, 178, 2019
|
|
6ZQR
 
 | |
5VUR
 
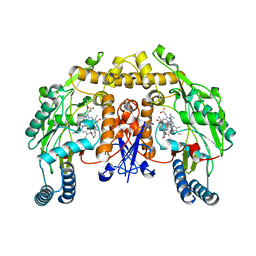 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 4-(2-(((2-Aminoquinolin-7-yl)methyl)amino)ethyl)-2-methylbenzonitrile | | Descriptor: | 4-(2-{[(2-aminoquinolin-7-yl)methyl]amino}ethyl)-2-methylbenzonitrile, 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-05-19 | | Release date: | 2017-08-16 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Hydrophilic, Potent, and Selective 7-Substituted 2-Aminoquinolines as Improved Human Neuronal Nitric Oxide Synthase Inhibitors.
J. Med. Chem., 60, 2017
|
|
6PPH
 
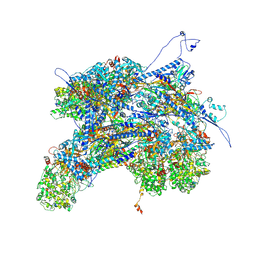 | | Kaposi's sarcoma-associated herpesvirus (KSHV), C1 penton vertex register, CATC-binding structure | | Descriptor: | Capsid vertex component 1, Capsid vertex component 2, Large tegument protein deneddylase, ... | | Authors: | Gong, D, Dai, X, Jih, J, Liu, Y.T, Bi, G.Q, Sun, R, Zhou, Z.H. | | Deposit date: | 2019-07-06 | | Release date: | 2019-09-11 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | DNA-Packing Portal and Capsid-Associated Tegument Complexes in the Tumor Herpesvirus KSHV.
Cell, 178, 2019
|
|
