1PLW
 
 | |
1PX3
 
 | | E. COLI (LACZ) BETA-GALACTOSIDASE (G794A) | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Juers, D.H, Hakda, S, Matthews, B.W, Huber, R.E. | | Deposit date: | 2003-07-02 | | Release date: | 2004-06-15 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Basis for the Altered Activity of Gly794 Variants of Escherichia coli Beta-Galactosidase
Biochemistry, 42, 2003
|
|
1S5L
 
 | | Architecture of the photosynthetic oxygen evolving center | | Descriptor: | 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, BETA-CAROTENE, BICARBONATE ION, ... | | Authors: | Ferreira, K.N, Iverson, T.M, Maghlaoui, K, Barber, J, Iwata, S. | | Deposit date: | 2004-01-21 | | Release date: | 2004-02-24 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Architecture of the Photosynthetic Oxygen-Evolving Center
Science, 303, 2004
|
|
1T36
 
 | | Crystal structure of E. coli carbamoyl phosphate synthetase small subunit mutant C248D complexed with uridine 5'-monophosphate | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, Carbamoyl-phosphate synthase large chain, ... | | Authors: | Thoden, J.B, Huang, X, Raushel, F.M, Holden, H.M. | | Deposit date: | 2004-04-24 | | Release date: | 2004-09-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Long-range allosteric transitions in carbamoyl phosphate synthetase.
Protein Sci., 13, 2004
|
|
5JXJ
 
 | | Structure of the proprotein convertase furin complexed to meta-guanidinomethyl-Phac-RVR-Amba in presence of EDTA | | Descriptor: | 2UC-ARG-VAL-ARG-00S, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JXI
 
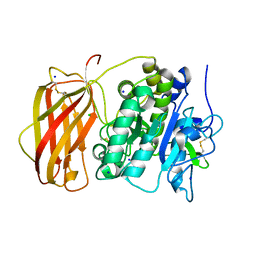 | | Structure of the unliganded form of the proprotein convertase furin in presence of EDTA. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Furin, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5JXG
 
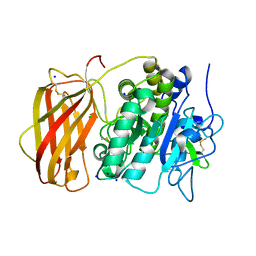 | | Structure of the unliganded form of the proprotein convertase furin. | | Descriptor: | CALCIUM ION, CHLORIDE ION, Furin, ... | | Authors: | Dahms, S.O, Arciniega, M, Steinmetzer, T, Huber, R, Than, M.E. | | Deposit date: | 2016-05-13 | | Release date: | 2016-10-05 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the unliganded form of the proprotein convertase furin suggests activation by a substrate-induced mechanism.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5VYW
 
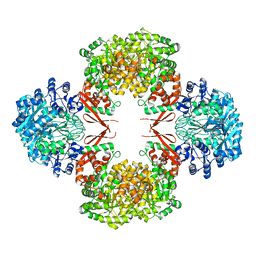 | | Crystal structure of Lactococcus lactis pyruvate carboxylase | | Descriptor: | BIOTIN, MANGANESE (II) ION, Pyruvate carboxylase | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2017-05-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VZ0
 
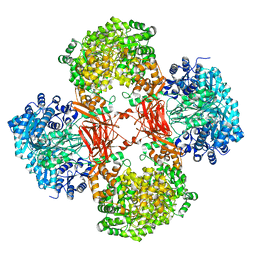 | | Crystal structure of Lactococcus lactis pyruvate carboxylase G746A mutant in complex with cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2017-05-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5VYZ
 
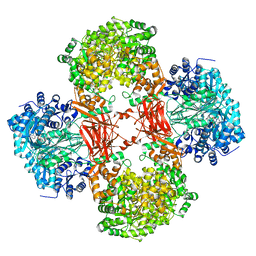 | | Crystal structure of Lactococcus lactis pyruvate carboxylase in complex with cyclic-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Choi, P.H, Tong, L. | | Deposit date: | 2017-05-26 | | Release date: | 2017-08-16 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural and functional studies of pyruvate carboxylase regulation by cyclic di-AMP in lactic acid bacteria.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
3A68
 
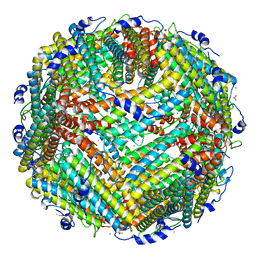 | | Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin | | Descriptor: | ACETIC ACID, CALCIUM ION, Ferritin-4, ... | | Authors: | Masuda, T, Goto, F, Yoshihara, T, Mikami, B. | | Deposit date: | 2009-08-26 | | Release date: | 2009-12-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of plant ferritin reveals a novel metal binding site that functions as a transit site for metal transfer in ferritin
J.Biol.Chem., 285, 2010
|
|
3T1Y
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA Lysine 3 (TRNALYS3) bound to an mRNA with an AAG-codon in the A-site and paromomycin | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Vendeix, F.A.P, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-25 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism.
J.Mol.Biol., 416, 2012
|
|
3T0D
 
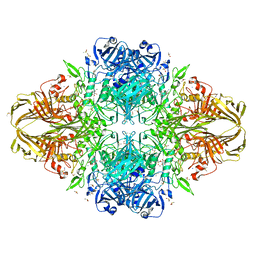 | | E.coli (lacZ) beta-galactosidase (S796T) in complex with galactonolactone | | Descriptor: | Beta-galactosidase, D-galactonolactone, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-20 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Ser-796 of Beta-galactosidase (E. coli) plays a Key role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T6E
 
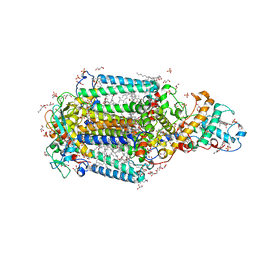 | | Crystal Structure of the Reaction Centre from Blastochloris viridis strain DSM 133 (ATCC 19567) substrain-94 | | Descriptor: | 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, BACTERIOPHEOPHYTIN B, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | New insights into the structure of the reaction centre from Blastochloris viridis: evolution in the laboratory.
Biochem.J., 442, 2012
|
|
3VD9
 
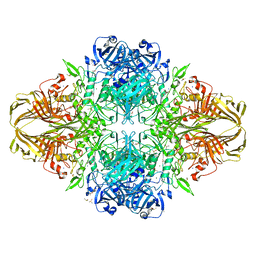 | | E. coli (lacZ) beta-galactosidase (N460S) in complex with IPTG | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Wheatley, R.W, Kappelhoff, J.C, Hahn, J.N, Dugdale, M.L, Dutkoski, M.J, Tamman, S.D, Fraser, M.E, Huber, R.E. | | Deposit date: | 2012-01-04 | | Release date: | 2012-04-11 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Substitution for Asn460 cripples {beta}-galactosidase (Escherichia coli) by increasing substrate affinity and decreasing transition state stability.
Arch.Biochem.Biophys., 521, 2012
|
|
2EIJ
 
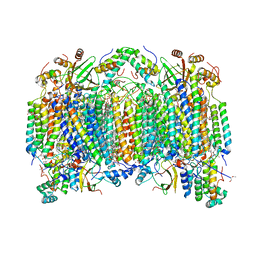 | | Bovine heart cytochrome C oxidase in the fully reduced state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Hirata, K, Shinzawa-Itoh, K, Yoko-o, S, Yamashita, E, Aoyama, H, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2007-03-13 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A histidine residue acting as a controlling site for dioxygen reduction and proton pumping by cytochrome c oxidase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3AG1
 
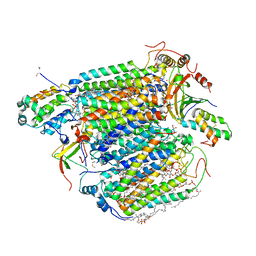 | | Bovine Heart Cytochrome c Oxidase in the Carbon Monoxide-bound Fully Reduced State at 280 K | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Ohta, K, Shinzawa-Itoh, K, Kanda, K, Taniguchi, M, Nabekura, H, Yamashita, E, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2010-03-19 | | Release date: | 2010-04-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Bovine cytochrome c oxidase structures enable O2 reduction with minimization of reactive oxygens and provide a proton-pumping gate
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
2ZZS
 
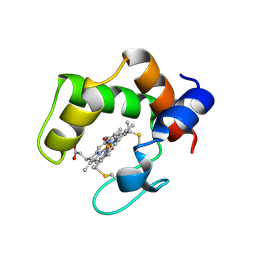 | | Crystal structure of cytochrome c554 from Vibrio parahaemolyticus strain RIMD2210633 | | Descriptor: | Cytochrome c554, GLYCEROL, HEME C | | Authors: | Akazaki, H, Kawai, F, Kumaki, Y, Sekine, K, Hakamata, W, Nishio, T, Park, S.-Y, Oku, T. | | Deposit date: | 2009-02-24 | | Release date: | 2010-03-16 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of cytochrome c554 from Vibrio parahaemolyticus strain RIMD2210633
To be Published
|
|
2EIM
 
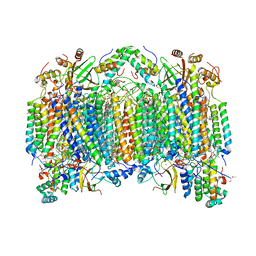 | | Zinc ion binding structure of bovine heart cytochrome C oxidase in the fully reduced state | | Descriptor: | (1R)-2-{[{[(2S)-2,3-DIHYDROXYPROPYL]OXY}(HYDROXY)PHOSPHORYL]OXY}-1-[(PALMITOYLOXY)METHYL]ETHYL (11E)-OCTADEC-11-ENOATE, (1S)-2-{[(2-AMINOETHOXY)(HYDROXY)PHOSPHORYL]OXY}-1-[(STEAROYLOXY)METHYL]ETHYL (5E,8E,11E,14E)-ICOSA-5,8,11,14-TETRAENOATE, (7R,17E,20E)-4-HYDROXY-N,N,N-TRIMETHYL-9-OXO-7-[(PALMITOYLOXY)METHYL]-3,5,8-TRIOXA-4-PHOSPHAHEXACOSA-17,20-DIEN-1-AMINIUM 4-OXIDE, ... | | Authors: | Muramoto, K, Hirata, K, Shinzawa-Itoh, K, Yoko-o, S, Yamashita, E, Aoyama, H, Tsukihara, T, Yoshikawa, S. | | Deposit date: | 2007-03-13 | | Release date: | 2007-05-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A histidine residue acting as a controlling site for dioxygen reduction and proton pumping by cytochrome c oxidase
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3T1H
 
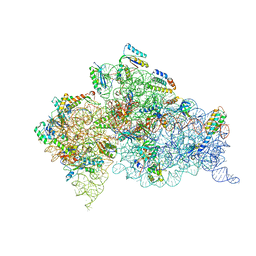 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a human anti-codon stem loop (HASL) of transfer RNA lysine 3 (tRNALys3) bound to an mRNA with an AAA-codon in the A-site and Paromomycin | | Descriptor: | 16s rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Murphy, F.V, Vendeix, F.A.P, Cantara, W, Leszczynska, G, Gustilo, E.M, Sproat, B, Malkiewicz, A.A.P, Agris, P.F. | | Deposit date: | 2011-07-21 | | Release date: | 2012-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Human tRNA(Lys3)(UUU) Is Pre-Structured by Natural Modifications for Cognate and Wobble Codon Binding through Keto-Enol Tautomerism.
J.Mol.Biol., 416, 2012
|
|
3T6D
 
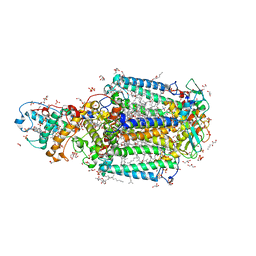 | | Crystal Structure of the Reaction Centre from Blastochloris viridis strain DSM 133 (ATCC 19567) substrain-08 | | Descriptor: | (2S,3R)-heptane-1,2,3-triol, 15-cis-1,2-dihydroneurosporene, BACTERIOCHLOROPHYLL B, ... | | Authors: | Roszak, A.W, Gardiner, A.T, Isaacs, N.W, Cogdell, R.J. | | Deposit date: | 2011-07-28 | | Release date: | 2011-11-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | New insights into the structure of the reaction centre from Blastochloris viridis: evolution in the laboratory.
Biochem.J., 442, 2012
|
|
3T8W
 
 | | A bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases | | Descriptor: | CARBONATE ION, M17 leucyl aminopeptidase, N-((2R,3S,6S,18S,21S)-2-amino-18-(4-benzoylbenzyl)-21-carbamoyl-3-hydroxy-6-(naphthalen-2-ylmethyl)-4,7,16,19-tetraoxo-1-phenyl-11,14-dioxa-5,8,17,20-tetraazapentacosan-25-yl)hex-5-ynamide, ... | | Authors: | McGowan, S, Klemba, M, Greebaum, D.C. | | Deposit date: | 2011-08-01 | | Release date: | 2011-09-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Bestatin-based chemical biology strategy reveals distinct roles for malaria M1- and M17-family aminopeptidases
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3BBX
 
 | |
3T0B
 
 | | E. coli (LacZ) beta-galactosidase (S796T) IPTG complex | | Descriptor: | 1-methylethyl 1-thio-beta-D-galactopyranoside, Beta-galactosidase, DIMETHYL SULFOXIDE, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-19 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | er-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
3T2O
 
 | | E. coli (lacZ) beta-galactosidase (S796D) | | Descriptor: | Beta-galactosidase, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Jancewicz, L.J, Wheatley, R.W, Sutendra, G, Lee, M, Fraser, M, Huber, R.E. | | Deposit date: | 2011-07-22 | | Release date: | 2012-01-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Ser-796 of Beta-Galactosidase (E. coli) Plays a Key Role in Maintaining an Optimum Balance between the Opened and Closed Conformations of the Catalytically Important Active Site Loop
Arch.Biochem.Biophys., 517, 2012
|
|
