7CRI
 
 | | 1 ps Structure of Chloride ion pumping rhodopsin (ClR) with NTQ motif | | Descriptor: | CHLORIDE ION, Chloride pumping rhodopsin, OLEIC ACID, ... | | Authors: | Yun, J.H, Liu, H, Lee, W.T, Schmidt, M. | | Deposit date: | 2020-08-13 | | Release date: | 2020-09-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Early-stage dynamics of chloride ion-pumping rhodopsin revealed by a femtosecond X-ray laser.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6KAF
 
 | | C2S2M2N2-type PSII-LHCII | | Descriptor: | (1R,3R)-6-{(3E,5E,7E,9E,11E,13E,15E,17E)-18-[(1S,4R,6R)-4-HYDROXY-2,2,6-TRIMETHYL-7-OXABICYCLO[4.1.0]HEPT-1-YL]-3,7,12,16-TETRAMETHYLOCTADECA-1,3,5,7,9,11,13,15,17-NONAENYLIDENE}-1,5,5-TRIMETHYLCYCLOHEXANE-1,3-DIOL, (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, (3S,5R,6S,3'S,5'R,6'S)-5,6,5',6'-DIEPOXY-5,6,5',6'- TETRAHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, ... | | Authors: | Chang, S.H, Shen, L.L, Huang, Z.H, Wang, W.D, Han, G.Y, Shen, J.R, Zhang, X. | | Deposit date: | 2019-06-22 | | Release date: | 2019-10-23 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.73 Å) | | Cite: | Structure of a C2S2M2N2-type PSII-LHCII supercomplex from the green algaChlamydomonas reinhardtii.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5ONX
 
 | | Resting state copper nitrite reductase determined by serial femtosecond rotation crystallography | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Halsted, T.P, Yamashita, K, Hirata, K, Ago, H, Ueno, G, Tosha, T, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An unprecedented dioxygen species revealed by serial femtosecond rotation crystallography in copper nitrite reductase.
IUCrJ, 5, 2018
|
|
5ONY
 
 | | As-isolated resting state copper nitrite reductase from Achromobacter xylosoxidans | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, COPPER (II) ION, Copper-containing nitrite reductase, ... | | Authors: | Halsted, T.P, Yamashita, K, Hirata, K, Ago, H, Ueno, G, Tosha, T, Eady, R.R, Antonyuk, S.V, Yamamoto, M, Hasnain, S.S. | | Deposit date: | 2017-08-04 | | Release date: | 2017-11-29 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An unprecedented dioxygen species revealed by serial femtosecond rotation crystallography in copper nitrite reductase.
IUCrJ, 5, 2018
|
|
9MQ4
 
 | | Damaged 70S ribosome with PrfH bound | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Tian, Y, Li, Q, Jin, H, Fatma, S, Zeng, F, Huang, R.H. | | Deposit date: | 2025-01-01 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.78 Å) | | Cite: | Molecular and structural basis of a subfamily of PrfH rescuing both the damaged and intact ribosomes stalled in translation.
Biorxiv, 2025
|
|
9MOR
 
 | | Damaged 70S ribosome with PrfH bound | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S11, ... | | Authors: | Tian, Y, Li, Q, Jin, H, Fatma, S, Zeng, F, Huang, R.H. | | Deposit date: | 2024-12-27 | | Release date: | 2025-02-05 | | Last modified: | 2025-03-19 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Molecular and structural basis of a subfamily of PrfH rescuing both the damaged and intact ribosomes stalled in translation.
Biorxiv, 2025
|
|
5JC9
 
 | | Structure of the Escherichia coli ribosome with the U1052G mutation in the 16S rRNA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-14 | | Release date: | 2016-07-06 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5J7L
 
 | | Structure of the 70S E coli ribosome with the U1052G mutation in the 16S rRNA bound to tetracycline | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 1,2-ETHANEDIOL, 1,4-DIAMINOBUTANE, ... | | Authors: | Cocozaki, A, Ferguson, A. | | Deposit date: | 2016-04-06 | | Release date: | 2016-07-27 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Resistance mutations generate divergent antibiotic susceptibility profiles against translation inhibitors.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6SRQ
 
 | |
6SRK
 
 | |
6SRO
 
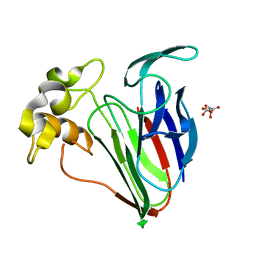 | |
6SRL
 
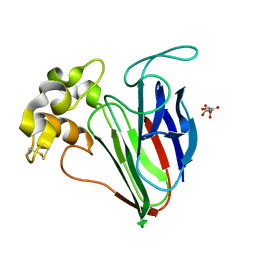 | |
6SRP
 
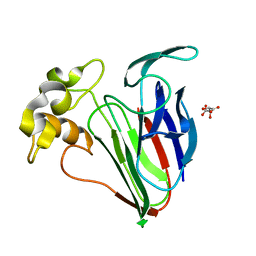 | |
4V7P
 
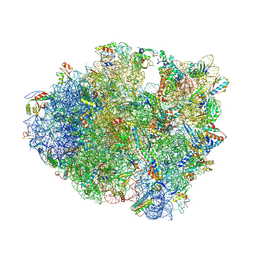 | | Recognition of the amber stop codon by release factor RF1. | | Descriptor: | 16S rRNA (1504-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Korostelev, A, Zhu, J, Asahara, H, Noller, H.F. | | Deposit date: | 2010-04-29 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (3.62 Å) | | Cite: | Recognition of the amber UAG stop codon by release factor RF1.
Embo J., 29, 2010
|
|
4V9N
 
 | | Crystal structure of the 70S ribosome bound with the Q253P mutant of release factor RF2. | | Descriptor: | 16S rRNA (1504-MER), 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Santos, N, Zhu, J, Donohue, J.P, Korostelev, A.A, Noller, H.F. | | Deposit date: | 2013-04-26 | | Release date: | 2014-07-09 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of the 70S Ribosome Bound with the Q253P Mutant Form of Release Factor RF2.
Structure, 21, 2013
|
|
4V7S
 
 | | Crystal structure of the E. coli ribosome bound to telithromycin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Dunkle, J.A, Xiong, L, Mankin, A.S, Cate, J.H.D. | | Deposit date: | 2010-08-05 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2547 Å) | | Cite: | Structures of the Escherichia coli ribosome with antibiotics bound near the peptidyl transferase center explain spectra of drug action.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3J3Y
 
 | |
3J3Q
 
 | |
8B0X
 
 | | Translating 70S ribosome in the unrotated state (P and E, tRNAs) | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fromm, S.A, O'Connor, K.M, Purdy, M, Bhatt, P.R, Loughran, G, Atkins, J.F, Jomaa, A, Mattei, S. | | Deposit date: | 2022-09-08 | | Release date: | 2022-11-30 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (1.55 Å) | | Cite: | The translating bacterial ribosome at 1.55 angstrom resolution generated by cryo-EM imaging services.
Nat Commun, 14, 2023
|
|
9E2G
 
 | | Cryo-EM structure of 48 nm repeat of microtubule doublet from T. brucei flagellum | | Descriptor: | CCDC81 HU domain-containing protein, CMF34/CARP4, Calcium-binding protein, ... | | Authors: | Xia, X, Shimogawa, M.M, Wang, H, Liu, S, Wijono, A, Langousis, G, Kassem, A.M, Wohlschlegel, J.A, Hill, K, Zhou, Z.H. | | Deposit date: | 2024-10-22 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Trypanosome doublet microtubule structures reveal flagellum assembly and motility mechanisms.
Science, 387, 2025
|
|
9E5C
 
 | | Cryo-EM structure of 96 nm repeat of microtubule doublet from T. brucei flagellum | | Descriptor: | 33 kDa inner dynein arm light chain, axonemal, putative, ... | | Authors: | Xia, X, Shimogawa, M.M, Wang, H, Liu, S, Wijono, A, Langousis, G, Kassem, A.M, Wohlschlegel, J.A, Hill, K, Zhou, Z.H. | | Deposit date: | 2024-10-28 | | Release date: | 2025-03-12 | | Last modified: | 2025-03-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Trypanosome doublet microtubule structures reveal flagellum assembly and motility mechanisms.
Science, 387, 2025
|
|
9Q87
 
 | | Principles of ion binding to RNA inferred from the analysis of a 1.55 Angstrom resolution bacterial ribosome structure - Part I: Mg2+ | | Descriptor: | 16S rRNA, 23S rRNA, 5S rRNA, ... | | Authors: | Leonarski, f, Henning-Knechtel, A, Kirmizialtin, S, Ennifar, E, Auffinger, P. | | Deposit date: | 2025-02-23 | | Release date: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (1.55 Å) | | Cite: | Principles of ion binding to RNA inferred from the analysis of a 1.55 Angs. resolution bacterial ribosome structure - Part I: Mg2+.
Nucleic Acids Res, 2024
|
|
7Q4K
 
 | | Erythromycin-stalled Escherichia coli 70S ribosome with streptococcal MsrDL nascent chain | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Fostier, C.R, Ousalem, F, Soufari, H, Leroy, E.C, Ngo, S, Innis, A, Hashem, Y, Boel, G. | | Deposit date: | 2021-10-31 | | Release date: | 2022-11-16 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Regulation of the macrolide resistance ABC-F translation factor MsrD.
Nat Commun, 14, 2023
|
|
8UVS
 
 | | Crystal structure of the wild-type Thermus thermophilus 70S ribosome in complex with spectinomycin derivative 2694, mRNA, deacylated A- and E-site tRNAphe, and deacylated P-site tRNAmet at 2.75A resolution | | Descriptor: | (2R,4R,4aS,5aR,6S,7S,8R,9S,9aR,10aS)-2-methyl-6,8-bis(methylamino)-4-({[2-(oxan-4-yl)ethyl]amino}methyl)octahydro-2H-pyrano[2,3-b][1,4]benzodioxine-4,4a,7,9(10aH)-tetrol, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Killam, B.Y, Phelps, G.A, Lee, R.E, Polikanov, Y.S. | | Deposit date: | 2023-11-03 | | Release date: | 2024-08-07 | | Last modified: | 2025-03-19 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Development of 2nd generation aminomethyl spectinomycins that overcome native efflux in Mycobacterium abscessus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
6MJZ
 
 | | Cryo-EM structure of Human Parainfluenza Virus Type 3 (hPIV3) in complex with antibody PIA174 | | Descriptor: | Fusion glycoprotein F0, PIA174 Fab Heavy chain, PIA174 Fab Light chain | | Authors: | Acharya, P, Stewart-Jones, G, Carragher, B, Potter, C.S, Kwong, P.D. | | Deposit date: | 2018-09-24 | | Release date: | 2018-11-14 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure-based design of a quadrivalent fusion glycoprotein vaccine for human parainfluenza virus types 1-4.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
