7D5A
 
 | | Crystal Structure of BACE1 in complex with N-{3-[(9S)-7-amino-2,2-difluoro-9-(prop-1-yn-1-yl)-6-oxa-8-azaspiro[3.5]non-7-en-9-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 1, GLYCEROL, IODIDE ION, ... | | Authors: | Fujimoto, K, Yoshida, S, Tadano, G, Asada, N, Fuchino, K, Suzuki, S, Matsuoka, E, Yamamoto, T, Yamamoto, S, Ando, S, Kanegawa, N, Tonomura, Y, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2020-09-25 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure-Based Approaches to Improving Selectivity through Utilizing Explicit Water Molecules: Discovery of Selective beta-Secretase (BACE1) Inhibitors over BACE2.
J.Med.Chem., 64, 2021
|
|
7D5U
 
 | | BACE2 xaperone complex with N-{3-[(9S)-7-amino-2,2-difluoro-9-(prop-1-yn-1-yl)-6-oxa-8-azaspiro[3.5]non-7-en-9-yl]-4-fluorophenyl}-5-cyanopyridine-2-carboxamide | | Descriptor: | Beta-secretase 2, N-[3-[(9S)-7-azanyl-2,2-bis(fluoranyl)-9-prop-1-ynyl-6-oxa-8-azaspiro[3.5]non-7-en-9-yl]-4-fluoranyl-phenyl]-5-cyano-pyridine-2-carboxamide, xaperone | | Authors: | Fujimoto, K, Yoshida, S, Tadano, G, Asada, N, Fuchino, K, Suzuki, S, Matsuoka, E, Yamamoto, T, Yamamoto, S, Ando, S, Kanegawa, N, Tonomura, Y, Ito, H, Moechars, D, Rombouts, F.J.R, Gijsen, H.J.M, Kusakabe, K.I. | | Deposit date: | 2020-09-28 | | Release date: | 2021-03-31 | | Last modified: | 2021-04-07 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure-Based Approaches to Improving Selectivity through Utilizing Explicit Water Molecules: Discovery of Selective beta-Secretase (BACE1) Inhibitors over BACE2.
J.Med.Chem., 64, 2021
|
|
5TM8
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC compound, 7-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)heptanoic acid | | Descriptor: | 7-{4-[(1S,4S,6R)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}heptanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Erumbi, R, Srinivasan, S, Bruno, N.E, Nowak, J, Izard, T, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM4
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC Analog, 5-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenoxy)pentanoic acid | | Descriptor: | 5-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenoxy}pentanoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TMM
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC analog, (E)-6-(4-((1R,4S,6R)-6-((4-bromophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)hex-5-enoic acid | | Descriptor: | 6-{4-[(1S,4S,6S)-6-[(4-bromophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}hex-5-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-13 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5TM9
 
 | | Crystal Structure of the ER-alpha Ligand-binding Domain (Y537S) in Complex with the OBHS-ASC Analog, (E)-3-(4-((1R,4S,6R)-6-((3-chlorophenoxy)sulfonyl)-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl)phenyl)acrylic acid | | Descriptor: | 3-{4-[(1S,4S,6R)-6-[(3-chlorophenoxy)sulfonyl]-3-(4-hydroxyphenyl)-7-oxabicyclo[2.2.1]hept-2-en-2-yl]phenyl}prop-2-enoic acid, Estrogen receptor, Nuclear receptor coactivator 2 | | Authors: | Nwachukwu, J.C, Srinivasan, S, Bruno, N.E, Nowak, J, Kojetin, D.J, Elemento, O, Katzenellenbogen, J.A, Nettles, K.W. | | Deposit date: | 2016-10-12 | | Release date: | 2017-01-18 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Systems Structural Biology Analysis of Ligand Effects on ER alpha Predicts Cellular Response to Environmental Estrogens and Anti-hormone Therapies.
Cell Chem Biol, 24, 2017
|
|
5CXI
 
 | | Crystal structure of Mycobacterium tuberculosis KstR in complex with 3-oxo-23,24-bisnorchol-4-en-22-oyl-CoA (4-BNC-CoA) | | Descriptor: | 3-oxo-23,24-bisnorchol-4-en-22-oyl-CoA, HTH-type transcriptional repressor KstR | | Authors: | Ho, N.A.T, Dawes, S, Kendall, S, Casabon, I, Crowe, A.M, Baker, E.N, Eltis, L.D, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-29 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
7LOB
 
 | | T4 lysozyme mutant L99A in complex with 1-fluoro-2-[(prop-2-en-1-yl)oxy]benzene | | Descriptor: | 1-fluoro-2-[(prop-2-en-1-yl)oxy]benzene, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, BETA-MERCAPTOETHANOL, ... | | Authors: | Kamenik, A.S, Singh, I, Lak, P, Balius, T.E, Liedl, K.R, Shoichet, B.K. | | Deposit date: | 2021-02-09 | | Release date: | 2021-05-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Energy penalties enhance flexible receptor docking in a model cavity.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
5JVD
 
 | | Tubulin-TUB092 complex | | Descriptor: | (2E)-3-(3-hydroxy-4-methoxyphenyl)-1-(7-methoxy-2H-1,3-benzodioxol-5-yl)-2-methylprop-2-en-1-one, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Canela, M.-D, Noppen, S, Bueno, O, Prota, A.E, Bargsten, K, Saez-Calvo, G, Jimeno, M.-L, Benkheil, M, Ribatti, D, Velazquez, S, Camarasa, M.-J, Diaz, J.F, Steinmetz, M.O, Priego, E.-M, Perez-Perez, M.-J, Liekens, S. | | Deposit date: | 2016-05-11 | | Release date: | 2016-06-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.393 Å) | | Cite: | Antivascular and antitumor properties of the tubulin-binding chalcone TUB091.
Oncotarget, 8, 2017
|
|
8GDI
 
 | | X-ray crystal structure of CYP124A1 from Mycobacterium Marinum in complex with 7-ketocholesterol | | Descriptor: | (3beta,8alpha,9beta)-3-hydroxycholest-5-en-7-one, Cytochrome P450 124A1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Ghith, A, Bruning, J.B, Bell, S.G. | | Deposit date: | 2023-03-06 | | Release date: | 2023-05-24 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | The oxidation of cholesterol derivatives by the CYP124 and CYP142 enzymes from Mycobacterium marinum.
J.Steroid Biochem.Mol.Biol., 231, 2023
|
|
8AM6
 
 | |
3PLZ
 
 | | Human LRH1 LBD bound to GR470 | | Descriptor: | (3aS,6aR)-5-[(4E)-oct-4-en-4-yl]-N,4-diphenyl-2,3,6,6a-tetrahydropentalen-3a(1H)-amine, 1,2-ETHANEDIOL, FTZ-F1 related protein, ... | | Authors: | Williams, S.P, Xu, R, Zuercher, W.J. | | Deposit date: | 2010-11-15 | | Release date: | 2011-03-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Small Molecule Agonists of the Orphan Nuclear Receptors Steroidogenic Factor-1 (SF-1, NR5A1) and Liver Receptor Homologue-1 (LRH-1, NR5A2).
J.Med.Chem., 54, 2011
|
|
8UN5
 
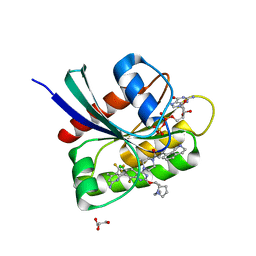 | | KRAS-G13D-GDP in complex with Cpd38 ((E)-1-((3S)-4-(7-(6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl)-6-chloro-8-fluoro-2-(((S)-2-methylenetetrahydro-1H-pyrrolizin-7a(5H)-yl)methoxy)quinazolin-4-yl)-3-methylpiperazin-1-yl)-3-(1,2,3,4-tetrahydroisoquinolin-8-yl)prop-2-en-1-one) | | Descriptor: | (2E)-1-{(3S)-4-[(7M)-7-[6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl]-6-chloro-8-fluoro-2-{[(4R,7aS)-2-methylidenetetrahydro-1H-pyrrolizin-7a(5H)-yl]methoxy}quinazolin-4-yl]-3-methylpiperazin-1-yl}-3-(1,2,3,4-tetrahydroisoquinolin-8-yl)prop-2-en-1-one, GLYCEROL, GTPase KRas, ... | | Authors: | Ultsch, M.H. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structure-Based Design and Evaluation of Reversible KRAS G13D Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8UN3
 
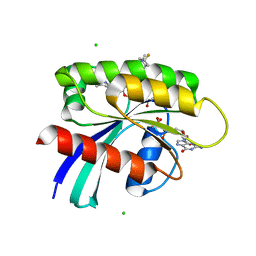 | | KRAS-G13D-GDP in complex with Cpd5 (1-((S)-10-(6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl)-11-chloro-7-(((2S,4R)-4-fluoro-1-methylpyrrolidin-2-yl)methoxy)-3,4,13,13a-tetrahydropyrazino[2',1':3,4][1,4]oxazepino[5,6,7-de]quinazolin-2(1H)-yl)prop-2-en-1-one) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(5M,8aS,13R)-5-[6-amino-4-methyl-3-(trifluoromethyl)pyridin-2-yl]-6-chloro-2-{[(2S,4R)-4-fluoro-1-methylpyrrolidin-2-yl]methoxy}-8a,9,11,12-tetrahydropyrazino[2',1':3,4][1,4]oxazepino[5,6,7-de]quinazolin-10(8H)-yl]prop-2-en-1-one, CHLORIDE ION, ... | | Authors: | Ultsch, M.H. | | Deposit date: | 2023-10-18 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Based Design and Evaluation of Reversible KRAS G13D Inhibitors.
Acs Med.Chem.Lett., 15, 2024
|
|
8UN4
 
 | |
8U0M
 
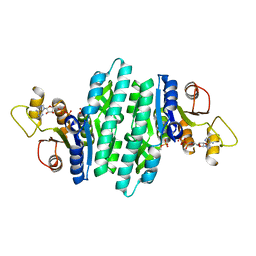 | | Crystal structure of isopentenyl phosphate kinase from Thermococcus paralvinellae bound to (E)-2-methylbut-2-en-1-yl monophosphate and ATP | | Descriptor: | (2E)-2-methylbut-2-en-1-yl dihydrogen phosphate, ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Singh, S, Thomas, L.M, Johnson, B.P, Brown, S. | | Deposit date: | 2023-08-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Ternary complexes of isopentenyl phosphate kinase from Thermococcus paralvinellae reveal molecular determinants of non-natural substrate specificity.
Proteins, 92, 2024
|
|
8U0L
 
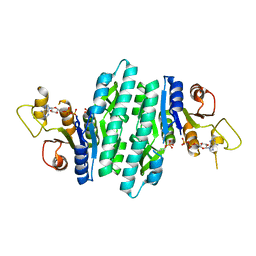 | | Crystal structure of isopentenyl phosphate kinase from Thermococcus paralvinellae bound to (E)-But-2-en-1-yl monophosphate and ADP | | Descriptor: | (2E)-but-2-en-1-yl dihydrogen phosphate, ADENOSINE-5'-DIPHOSPHATE, Isopentenyl phosphate kinase | | Authors: | Singh, S, Thomas, L.M, Johnson, B.P, Brown, S. | | Deposit date: | 2023-08-29 | | Release date: | 2024-02-21 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Ternary complexes of isopentenyl phosphate kinase from Thermococcus paralvinellae reveal molecular determinants of non-natural substrate specificity.
Proteins, 92, 2024
|
|
2KK2
 
 | | NMR solution structure of the pheromone En-A1 from Euplotes nobilii | | Descriptor: | En-A1 | | Authors: | Pedrini, B, Alimenti, C, Vallesi, A, Luporini, P, Wuthrich, K. | | Deposit date: | 2009-06-15 | | Release date: | 2010-05-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Antarctic and Arctic populations of the ciliate Euplotes nobilii show common pheromone-mediated cell-cell signaling and cross-mating.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3BPC
 
 | | co-crystal structure of S25-2 Fab in complex with 5-deoxy-4-epi-2,3-dehydro Kdo (4.8) Kdo | | Descriptor: | 3,4,5-trideoxy-alpha-D-erythro-oct-3-en-2-ulopyranosonic acid-(2-8)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, Fab, antibody fragment (IgG1k), ... | | Authors: | Brooks, C.L, Evans, S.V. | | Deposit date: | 2007-12-18 | | Release date: | 2009-01-13 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Exploration of specificity in germline monoclonal antibody recognition of a range of natural and synthetic epitopes.
J.Mol.Biol., 377, 2008
|
|
1W9X
 
 | | Bacillus halmapalus alpha amylase | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranose, ALPHA AMYLASE, CALCIUM ION, ... | | Authors: | Davies, G.J, Brzozowski, A.M, Dauter, Z, Rasmussen, M.D, Borchert, T.V, Wilson, K.S. | | Deposit date: | 2004-10-20 | | Release date: | 2005-02-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of a Bacillus Halmapalus Family 13 Alpha-Amylase, Bha, in Complex with an Acarbose-Derived Nonasaccharide at 2.1 A Resolution
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1MXG
 
 | | Crystal Structure of a (Ca,Zn)-dependent alpha-amylase from the hyperthermophilic archaeon Pyrococcus woesei in complex with acarbose | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-{2-[2-2-(METHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Linden, A, Mayans, O, Meyer-Klaucke, W, Antranikian, G, Wilmanns, M. | | Deposit date: | 2002-10-02 | | Release date: | 2003-06-10 | | Last modified: | 2020-09-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Differential Regulation of a Hyperthermophilic alpha-Amylase with a Novel (Ca,Zn) Two-metal Center by Zinc
J.Biol.Chem., 278, 2003
|
|
2R1W
 
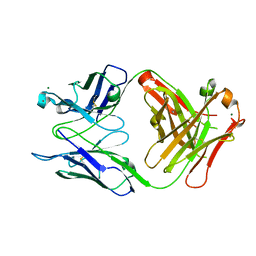 | | Crystal structure of S25-2 Fab in complex with Kdo analogues | | Descriptor: | 3,4,5-trideoxy-alpha-D-erythro-oct-3-en-2-ulopyranosonic acid-(2-8)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, Fab, antibody fragment (IgG1k), ... | | Authors: | Brooks, C.L, Evans, S.V. | | Deposit date: | 2007-08-23 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Exploration of specificity in germline monoclonal antibody recognition of a range of natural and synthetic epitopes.
J.Mol.Biol., 377, 2008
|
|
8WB4
 
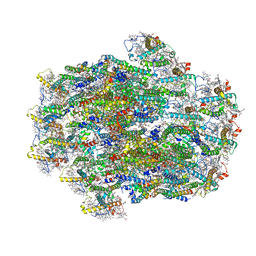 | | Structure of PSII-ACPII supercomplex from cryptophyte algae | | Descriptor: | (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E})-3,7,12,16-tetramethyl-18-[(4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohexen-1-yl]octadeca-3,5,7,9,11,13,15-heptaen-1,17-diynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-(2,6,6-trimethylcyclohexen-1-yl)octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, (1~{R})-3,5,5-trimethyl-4-[(3~{E},5~{E},7~{E},9~{E},11~{E},13~{E},15~{E},17~{E})-3,7,12,16-tetramethyl-18-[(1~{R},4~{R})-2,6,6-trimethyl-4-oxidanyl-cyclohex-2-en-1-yl]octadeca-3,5,7,9,11,13,15,17-octaen-1-ynyl]cyclohex-3-en-1-ol, ... | | Authors: | Li, X.Y, Mao, Z.Y, Shen, J.R, Han, G.Y. | | Deposit date: | 2023-09-08 | | Release date: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structure and distinct supramolecular organization of a PSII-ACPII dimer from a cryptophyte alga Chroomonas placoidea.
Nat Commun, 15, 2024
|
|
3TOP
 
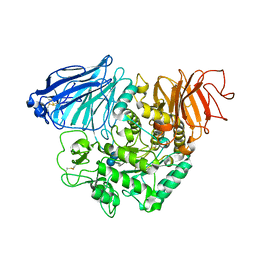 | | Crystral Structure of the C-terminal Subunit of Human Maltase-Glucoamylase in Complex with Acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, Maltase-glucoamylase, intestinal | | Authors: | Shen, Y, Qin, X.H, Ren, L.M. | | Deposit date: | 2011-09-06 | | Release date: | 2011-11-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.881 Å) | | Cite: | Structural insight into substrate specificity of human intestinal maltase-glucoamylase
Protein Cell, 2, 2011
|
|
2R23
 
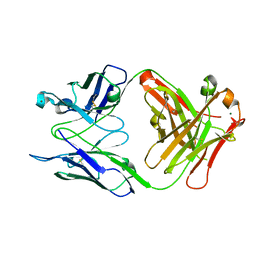 | | Crystal structure of S25-2 Fab in complex with Kdo analogues | | Descriptor: | D-glycero-alpha-D-talo-oct-2-ulopyranosonic acid-(2-4)-prop-2-en-1-yl 3-deoxy-alpha-D-manno-oct-2-ulopyranosidonic acid, Fab, antibody fragment (IgG1k), ... | | Authors: | Brooks, C.L, Evans, S.V. | | Deposit date: | 2007-08-24 | | Release date: | 2008-12-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Exploration of specificity in germline monoclonal antibody recognition of a range of natural and synthetic epitopes.
J.Mol.Biol., 377, 2008
|
|
