6ZY8
 
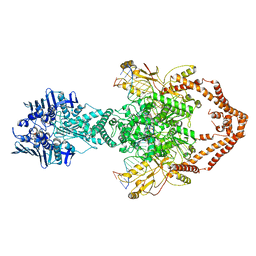 | | Cryo-EM structure of the entire Human topoisomerase II alpha in State 2 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
6ZY6
 
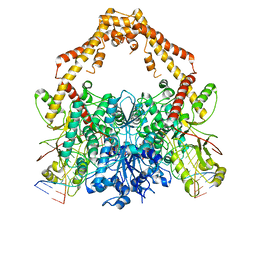 | | Cryo-EM structure of the Human topoisomerase II alpha DNA-binding/cleavage domain in State 2 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
6ZY5
 
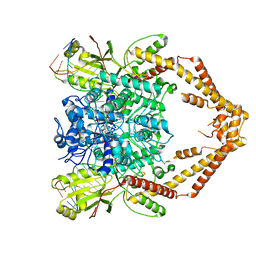 | | Cryo-EM structure of the Human topoisomerase II alpha DNA-binding/cleavage domain in State 1 | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, DNA (5'-D(*CP*GP*CP*GP*CP*AP*TP*CP*GP*TP*CP*AP*TP*CP*CP*TP*C)-3'), DNA (5'-D(*GP*AP*GP*GP*AP*TP*GP*AP*CP*GP*AP*TP*G)-3'), ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2020-07-30 | | Release date: | 2021-05-26 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural basis for allosteric regulation of Human Topoisomerase II alpha.
Nat Commun, 12, 2021
|
|
3EAY
 
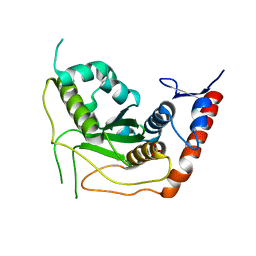 | | Crystal structure of the human SENP7 catalytic domain | | Descriptor: | SULFATE ION, Sentrin-specific protease 7 | | Authors: | Lima, C.D, Reverter, D. | | Deposit date: | 2008-08-26 | | Release date: | 2008-09-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of the Human SENP7 Catalytic Domain and Poly-SUMO Deconjugation Activities for SENP6 and SENP7.
J.Biol.Chem., 283, 2008
|
|
7YQ8
 
 | | Cryo-EM structure of human topoisomerase II beta in complex with DNA and etoposide | | Descriptor: | (5S,5aR,8aR,9R)-9-(4-hydroxy-3,5-dimethoxyphenyl)-8-oxo-5,5a,6,8,8a,9-hexahydrofuro[3',4':6,7]naphtho[2,3-d][1,3]dioxol -5-yl 4,6-O-[(1R)-ethylidene]-beta-D-glucopyranoside, 50-mer DNA, DNA topoisomerase 2-beta, ... | | Authors: | Naganuma, M, Ehara, H, Kim, D, Nakagawa, R, Cong, A, Bu, H, Jeong, J, Jang, J, Schellenberg, M.J, Bunch, H, Sekine, S. | | Deposit date: | 2022-08-05 | | Release date: | 2024-01-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | ERK2-topoisomerase II regulatory axis is important for gene activation in immediate early genes.
Nat Commun, 14, 2023
|
|
1WYW
 
 | | Crystal Structure of SUMO1-conjugated thymine DNA glycosylase | | Descriptor: | CHLORIDE ION, G/T mismatch-specific thymine DNA glycosylase, MAGNESIUM ION, ... | | Authors: | Baba, D, Maita, N, Jee, J.G, Uchimura, Y, Saitoh, H, Sugasawa, K, Hanaoka, F, Tochio, H, Hiroaki, H, Shirakawa, M. | | Deposit date: | 2005-02-17 | | Release date: | 2005-06-21 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of thymine DNA glycosylase conjugated to SUMO-1.
Nature, 435, 2005
|
|
2VXD
 
 | |
4W5V
 
 | | Crystal structure of Human SUMO E2-conjugating enzyme (Ubc9) in complex with E1-activating enzyme (Uba2) ubiquitin fold domain (Ufd) | | Descriptor: | FORMIC ACID, GLYCEROL, POTASSIUM ION, ... | | Authors: | Boucher, L.E, Reiter, K.H, Matunis, M.J, Bosch, J. | | Deposit date: | 2014-08-19 | | Release date: | 2015-09-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of human SUMO complex
To Be Published
|
|
4WJQ
 
 | | Crystal Structure of SUMO1 in complex with Daxx | | Descriptor: | Daxx, Small ubiquitin-related modifier 1 | | Authors: | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
4WJN
 
 | | Crystal structure of SUMO1 in complex with phosphorylated PML | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
4WJP
 
 | | Crystal Structure of SUMO1 in complex with phosphorylated Daxx | | Descriptor: | Daxx, Small ubiquitin-related modifier 1 | | Authors: | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
4WJO
 
 | | Crystal Structure of SUMO1 in complex with PML | | Descriptor: | Protein PML, Small ubiquitin-related modifier 1 | | Authors: | Cappadocia, L, Mascle, X.H, Bourdeau, V, Tremblay-Belzile, S, Chaker-Margot, M, Lussier-Price, M, Wada, J, Sakaguchi, K, Aubry, M, Ferbeyre, G, Omichinski, J.G. | | Deposit date: | 2014-10-01 | | Release date: | 2014-12-31 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Structural and Functional Characterization of the Phosphorylation-Dependent Interaction between PML and SUMO1.
Structure, 23, 2015
|
|
8HAL
 
 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 1 | | Descriptor: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAG
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (3.2 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAH
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (3.9 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAI
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 1 (4.7 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAM
 
 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 2 | | Descriptor: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAJ
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 2 (4.8 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAN
 
 | | Cryo-EM structure of the CBP catalytic core bound to the H4K12acK16ac nucleosome, class 3 | | Descriptor: | CREB-binding protein, DNA (180-mer), Histone H2A type 1-B/E, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
8HAK
 
 | | Cryo-EM structure of the p300 catalytic core bound to the H4K12acK16ac nucleosome, class 4 (4.5 angstrom resolution) | | Descriptor: | DNA (180-mer), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Kikuchi, M, Morita, S, Wakamori, M, Shin, S, Uchikubo-Kamo, T, Shirouzu, M, Umehara, T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Epigenetic mechanisms to propagate histone acetylation by p300/CBP.
Nat Commun, 14, 2023
|
|
7AAV
 
 | | Human pre-Bact-2 spliceosome core structure | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, Cell division cycle 5-like protein, D-chiro inositol hexakisphosphate, ... | | Authors: | Townsend, C, Kastner, B, Leelaram, M.N, Bertram, K, Stark, H, Luehrmann, R. | | Deposit date: | 2020-09-04 | | Release date: | 2020-12-09 | | Last modified: | 2020-12-30 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Mechanism of protein-guided folding of the active site U2/U6 RNA during spliceosome activation.
Science, 370, 2020
|
|
7AOA
 
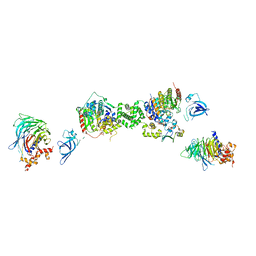 | | Structure of the extended MTA1/HDAC1/MBD2/RBBP4 NURD deacetylase complex | | Descriptor: | Histone deacetylase 1, Histone-binding protein RBBP4, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Millard, C.J, Fairall, L, Ragan, T.J, Savva, C.G, Schwabe, J.W.R. | | Deposit date: | 2020-10-14 | | Release date: | 2020-11-11 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (19.4 Å) | | Cite: | The topology of chromatin-binding domains in the NuRD deacetylase complex.
Nucleic Acids Res., 48, 2020
|
|
6ID0
 
 | | Cryo-EM structure of a human intron lariat spliceosome prior to Prp43 loaded (ILS1 complex) at 2.9 angstrom resolution | | Descriptor: | 116 kDa U5 small nuclear ribonucleoprotein component, CWF19-like protein 2, Cell division cycle 5-like protein, ... | | Authors: | Zhang, X, Zhan, X, Yan, C, Shi, Y. | | Deposit date: | 2018-09-07 | | Release date: | 2019-03-13 | | Last modified: | 2020-10-14 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structures of the human spliceosomes before and after release of the ligated exon.
Cell Res., 29, 2019
|
|
7DN3
 
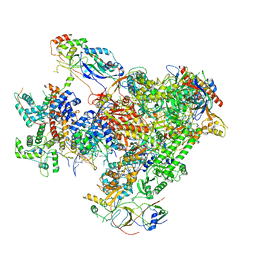 | | Structure of Human RNA Polymerase III elongation complex | | Descriptor: | DNA (5'-D(P*TP*CP*GP*TP*CP*TP*GP*AP*TP*CP*TP*CP*GP*GP*AP*A)-3'), DNA (5'-D(P*TP*TP*CP*CP*GP*AP*GP*AP*TP*CP*AP*GP*AP*CP*GP*AP*GP*AP*TP*CP*GP*GP*G)-3'), DNA-directed RNA polymerase III subunit RPC1, ... | | Authors: | Li, L, Yu, Z, Zhao, D, Ren, Y, Hou, H, Xu, Y. | | Deposit date: | 2020-12-08 | | Release date: | 2021-03-17 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of human RNA polymerase III elongation complex.
Cell Res., 31, 2021
|
|
1KPS
 
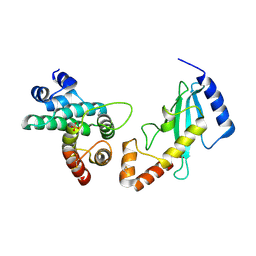 | | Structural Basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin conjugating enzyme Ubc9 and RanGAP1 | | Descriptor: | Ran-GTPase activating protein 1, SULFATE ION, Ubiquitin-like protein SUMO-1 conjugating enzyme | | Authors: | Bernier-Villamor, V, Sampson, D.A, Matunis, M.J, Lima, C.D. | | Deposit date: | 2002-01-02 | | Release date: | 2002-02-13 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for E2-mediated SUMO conjugation revealed by a complex between ubiquitin-conjugating enzyme Ubc9 and RanGAP1.
Cell(Cambridge,Mass.), 108, 2002
|
|
