3LQW
 
 | |
3MDX
 
 | |
3MBQ
 
 | |
3H6X
 
 | |
3LOJ
 
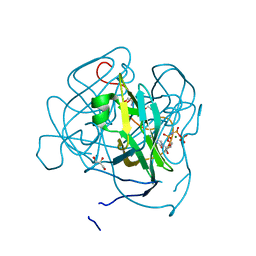 | | Structure of Mycobacterium tuberculosis dUTPase H145A mutant | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Deoxyuridine 5'-triphosphate nucleotidohydrolase, ... | | Authors: | Leveles, I, Harmat, V, Pecsi, I, Lopata, A, Vertessy, B.G, Toth, J. | | Deposit date: | 2010-02-04 | | Release date: | 2010-08-11 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Aromatic stacking between nucleobase and enzyme promotes phosphate ester hydrolysis in dUTPase
Nucleic Acids Res., 38, 2010
|
|
2XCE
 
 | | Structure of YncF in complex with dUpNHpp | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CALCIUM ION, GLYCEROL, ... | | Authors: | Garcia-Nafria, J, Burchell, L, Takezawa, M, Rzechorzek, N, Fogg, M, Wilson, K.S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of the Genomic Bacillus Subtilis Dutpase: Novel Features in the Phe-Lid.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2XCD
 
 | | Structure of YncF,the genomic dUTPase from Bacillus subtilis | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Garcia, J, Burchell, L, Takezawa, M, Rzechorzek, N.J, Fogg, M, Wilson, K.S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Structure of the Genomic Bacillus Subtilis Dutpase: Novel Features in the Phe-Lid.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
3P48
 
 | | Structure of the yeast dUTPase DUT1 in complex with dUMPNPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Petit, P, Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Yakunin, A.F, Savchenko, A, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-10-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
3ARA
 
 | | Discovery of Novel Uracil Derivatives as Potent Human dUTPase Inhibitors | | Descriptor: | 1-[3-({(2R)-2-[hydroxy(diphenyl)methyl]pyrrolidin-1-yl}sulfonyl)propyl]pyrimidine-2,4(1H,3H)-dione, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Chong, K.T, Miyakoshi, H, Miyahara, S, Fukuoka, M. | | Deposit date: | 2010-11-25 | | Release date: | 2010-12-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis and discovery of N-carbonylpyrrolidine- or N-sulfonylpyrrolidine-containing uracil derivatives as potent human deoxyuridine triphosphatase inhibitors
J.Med.Chem., 55, 2012
|
|
3ARN
 
 | | Human dUTPase in complex with novel uracil derivative | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION, N-{5-[(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)methoxy]-2-methylpentan-2-yl}benzenesulfonamide | | Authors: | Chong, K.T, Miyahara, S, Miyakoshi, H, Fukuoka, M. | | Deposit date: | 2010-12-03 | | Release date: | 2010-12-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of a novel class of potent human deoxyuridine triphosphatase inhibitors remarkably enhancing the antitumor activity of thymidylate synthase inhibitors
J.Med.Chem., 55, 2012
|
|
2Y1T
 
 | | Bacillus subtilis prophage dUTPase in complex with dUDP | | Descriptor: | DEOXYURIDINE-5'-DIPHOSPHATE, SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harkiolaki, M, Persson, R, Fogg, M.J, Wilson, K.S. | | Deposit date: | 2010-12-10 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | The Structure of Bacillus Subtilis Sp Beta Prophage Dutpase and its Complexes with Two Nucleotides
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XY3
 
 | | Structure of the Bacillus subtilis prophage dUTPase with dUpNHpp | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, MAGNESIUM ION, SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harkiolaki, M, Persson, R, Fogg, M.J, Wilson, K.S. | | Deposit date: | 2010-11-12 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | The Structure of Bacillus Subtilis Sp Beta Prophage Dutpase and its Complexes with Two Nucleotides
Acta Crystallogr.,Sect.D, 67, 2011
|
|
2XX6
 
 | | Structure of the Bacillus subtilis prophage dUTPase, YosS | | Descriptor: | SPBC2 PROPHAGE-DERIVED DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDOHYDROLASE YOSS | | Authors: | Garcia-Nafria, J, Harkiolaki, M, Persson, R, Fogg, M.J, Wilson, K.S. | | Deposit date: | 2010-11-08 | | Release date: | 2011-02-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | The Structure of Bacillus Subtilis Sp Beta Prophage Dutpase and its Complexes with Two Nucleotides
Acta Crystallogr.,Sect.D, 67, 2011
|
|
3TQZ
 
 | | Structure of a deoxyuridine 5'-triphosphate nucleotidohydrolase (dut) from Coxiella burnetii | | Descriptor: | Deoxyuridine 5'-triphosphate nucleotidohydrolase, SULFATE ION | | Authors: | Cheung, J, Franklin, M, Rudolph, M, Cassidy, M, Gary, E, Burshteyn, F, Love, J. | | Deposit date: | 2011-09-09 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural genomics for drug design against the pathogen Coxiella burnetii.
Proteins, 83, 2015
|
|
3TP1
 
 | |
3TPW
 
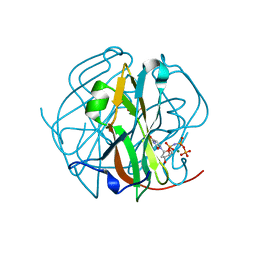 | | CRYSTAL STRUCTURE OF M-PMV DUTPASE - DUPNPP complex revealing distorted ligand geometry (approach intermediate) | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDO HYDROLASE, ... | | Authors: | Barabas, O, Nemeth, V, Vertessy, B.G. | | Deposit date: | 2011-09-08 | | Release date: | 2011-10-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Snapshots of Enzyme-Catalysed Phosphate Ester Hydrolysis Directly Visualize In-line Attack and Inversion
to be published
|
|
3TQ3
 
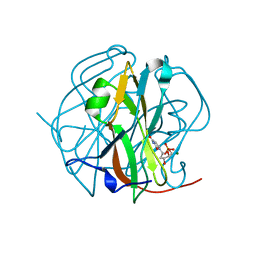 | |
3TRN
 
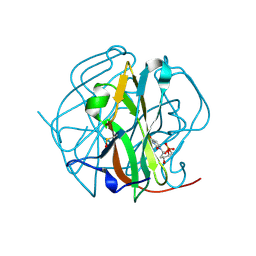 | |
3TPS
 
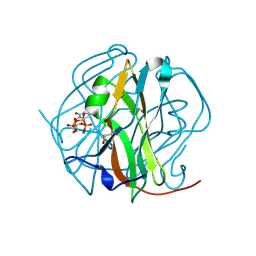 | | Crystal structure of M-PMV dUTPASE complexed with dUPNPP substrate | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DEOXYURIDINE 5'-TRIPHOSPHATE NUCLEOTIDO HYDROLASE, ... | | Authors: | Barabas, O, Nemeth, V, Vertessy, B.G. | | Deposit date: | 2011-09-08 | | Release date: | 2011-10-12 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Snapshots of Enzyme-Catalysed Phosphate Ester Hydrolysis Directly Visualize In-line Attack and Inversion
To be Published
|
|
3TPY
 
 | | Crystal structure of M-PMV dUTPase with a mixed population of substrate (dUPNPP) and post-inversion product (dUMP) in the active sites | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Barabas, O, Nemeth, V, Vertessy, B.G. | | Deposit date: | 2011-09-08 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Snapshots of Enzyme-Catalysed Phosphate Ester Hydrolysis Directly Visualize In-line Attack and Inversion
to be published
|
|
3TTA
 
 | |
3TSL
 
 | |
3TPN
 
 | | Crystal structure of M-PMV dUTPASE complexed with dUPNPP, substrate | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Nemeth, V, Barabas, O, Vertessy, G.B. | | Deposit date: | 2011-09-08 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Snapshots of Enzyme-Catalysed Phosphate Ester Hydrolysis Directly Visualize In-line Attack and Inversion
To be Published
|
|
3TQ4
 
 | | Crystal structure of M-PMV dUTPase with a mixed population of substrate (dUPNPP) and post-inversion product (dUMP) in the active sites | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, 2'-DEOXYURIDINE 5'-MONOPHOSPHATE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Barabas, O, Nemeth, V, Vertessy, B.G. | | Deposit date: | 2011-09-09 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Snapshots of Enzyme-Catalysed Phosphate Ester Hydrolysis Directly Visualize In-line Attack and Inversion
to be published
|
|
3TRL
 
 | |
