6XTY
 
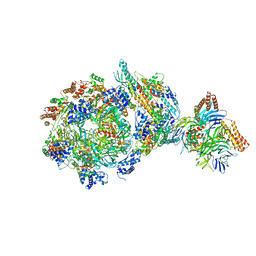 | | CryoEM structure of human CMG bound to AND-1 (CMGA) | | Descriptor: | Cell division control protein 45 homolog, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Rzechorzek, N.J, Pellegrini, L, Chirgadze, D.Y, Hardwick, S.W. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-15 | | Method: | ELECTRON MICROSCOPY (6.77 Å) | | Cite: | CryoEM structures of human CMG-ATP gamma S-DNA and CMG-AND-1 complexes.
Nucleic Acids Res., 48, 2020
|
|
6XTX
 
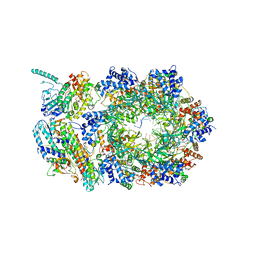 | | CryoEM structure of human CMG bound to ATPgammaS and DNA | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Cell division control protein 45 homolog, DNA (5'-D(P*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), ... | | Authors: | Rzechorzek, N.J, Pellegrini, L. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-15 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | CryoEM structures of human CMG-ATP gamma S-DNA and CMG-AND-1 complexes.
Nucleic Acids Res., 48, 2020
|
|
8QHQ
 
 | | Crystal structure of human DNPH1 bound to hmdUMP | | Descriptor: | 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, 5-HYDROXYMETHYLURIDINE-2'-DEOXY-5'-MONOPHOSPHATE | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
8QHR
 
 | | Crystal structure of the human DNPH1 glycosyl-enzyme intermediate | | Descriptor: | 1',2'-DIDEOXYRIBOFURANOSE-5'-PHOSPHATE, 1,2-ETHANEDIOL, 2'-deoxynucleoside 5'-phosphate N-hydrolase 1, ... | | Authors: | Rzechorzek, N.J, West, S.C. | | Deposit date: | 2023-09-09 | | Release date: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of substrate hydrolysis by the human nucleotide pool sanitiser DNPH1.
Nat Commun, 14, 2023
|
|
4D2I
 
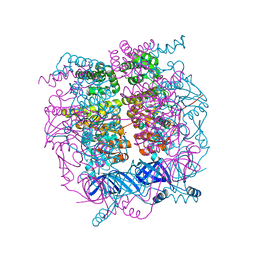 | | Crystal structure of the HerA hexameric DNA translocase from Sulfolobus solfataricus bound to AMP-PNP | | Descriptor: | HERA, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | Rzechorzek, N.J, Blackwood, J.K, Bray, S.M, Maman, J.D, Pellegrini, L, Robinson, N.P. | | Deposit date: | 2014-05-09 | | Release date: | 2014-12-03 | | Method: | X-RAY DIFFRACTION (2.841 Å) | | Cite: | Structure of the Hexameric Hera ATPase Reveals a Mechanism of Translocation-Coupled DNA-End Processing in Archaea
Nat.Commun., 5, 2014
|
|
2YGK
 
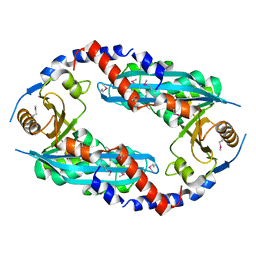 | | Crystal structure of the NurA nuclease from Sulfolobus solfataricus | | Descriptor: | MANGANESE (II) ION, NURA | | Authors: | Rzechorzek, N.J, Blackwood, J.K, Abrams, A.S, Maman, J.D, Robinson, N.P, Pellegrini, L. | | Deposit date: | 2011-04-18 | | Release date: | 2011-12-14 | | Last modified: | 2012-04-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural and Functional Insights Into DNA-End Processing by the Archaeal Hera Helicase-Nura Nuclease Complex.
Nucleic Acids Res., 40, 2012
|
|
3G65
 
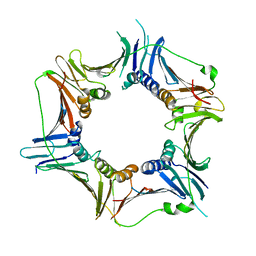 | | Crystal Structure of the Human Rad9-Rad1-Hus1 DNA Damage Checkpoint Complex | | Descriptor: | Cell cycle checkpoint control protein RAD9A, Cell cycle checkpoint protein RAD1, Checkpoint protein HUS1 | | Authors: | Dore, A.S, Kilkenny, M.L, Rzechorzek, N.J, Pearl, L.H. | | Deposit date: | 2009-02-06 | | Release date: | 2009-05-26 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of the rad9-rad1-hus1 DNA damage checkpoint complex--implications for clamp loading and regulation.
Mol.Cell, 34, 2009
|
|
2C0S
 
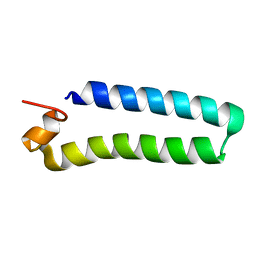 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-09-07 | | Release date: | 2006-09-25 | | Last modified: | 2018-05-09 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
2BZB
 
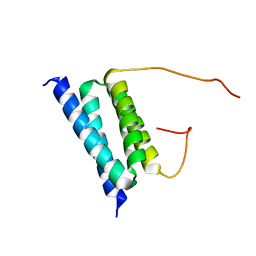 | | NMR Solution Structure of a protein aspartic acid phosphate phosphatase from Bacillus Anthracis | | Descriptor: | CONSERVED DOMAIN PROTEIN | | Authors: | Grenha, R, Rzechorzek, N.J, Brannigan, J.A, Ab, E, Folkers, G.E, De Jong, R.N, Diercks, T, Wilkinson, A.J, Kaptein, R, Wilson, K.S. | | Deposit date: | 2005-08-14 | | Release date: | 2006-09-25 | | Last modified: | 2018-05-09 | | Method: | SOLUTION NMR | | Cite: | Structural characterization of Spo0E-like protein-aspartic acid phosphatases that regulate sporulation in bacilli.
J. Biol. Chem., 281, 2006
|
|
2XCD
 
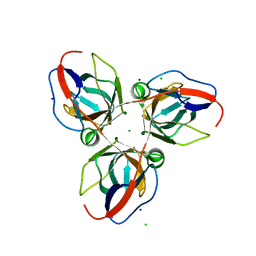 | | Structure of YncF,the genomic dUTPase from Bacillus subtilis | | Descriptor: | CHLORIDE ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Garcia, J, Burchell, L, Takezawa, M, Rzechorzek, N.J, Fogg, M, Wilson, K.S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | The Structure of the Genomic Bacillus Subtilis Dutpase: Novel Features in the Phe-Lid.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
2XCE
 
 | | Structure of YncF in complex with dUpNHpp | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CALCIUM ION, GLYCEROL, ... | | Authors: | Garcia-Nafria, J, Burchell, L, Takezawa, M, Rzechorzek, N, Fogg, M, Wilson, K.S. | | Deposit date: | 2010-04-22 | | Release date: | 2010-08-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The Structure of the Genomic Bacillus Subtilis Dutpase: Novel Features in the Phe-Lid.
Acta Crystallogr.,Sect.D, 66, 2010
|
|
