8TOF
 
 | | Rpd3S bound to an H3K36Cme3 modified nucleosome | | Descriptor: | Chromatin modification-related protein EAF3, DNA (176-MER), Histone H2A, ... | | Authors: | Markert, J.W, Vos, S.M, Farnung, L. | | Deposit date: | 2023-08-03 | | Release date: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structure of the complete Saccharomyces cerevisiae Rpd3S-nucleosome complex.
Nat Commun, 14, 2023
|
|
8TXW
 
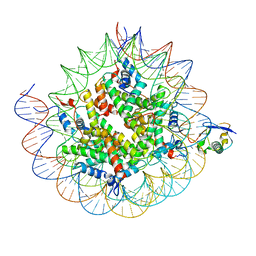 | | Cryo-EM structure of the human nucleosome core particle ubiquitylated at histone H2A K15 in complex with RNF168 (Class 2) | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-08-24 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQE
 
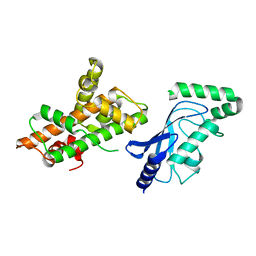 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 26-residue linker (RING not modeled in density) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.562 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQB
 
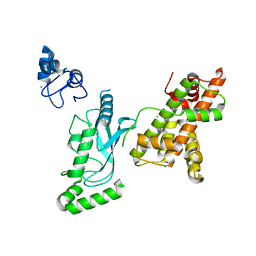 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 1) | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.484 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQA
 
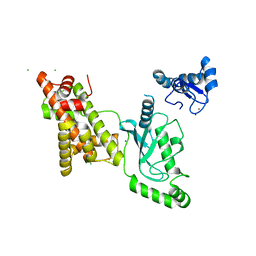 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 12-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, SODIUM ION, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.049 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UPF
 
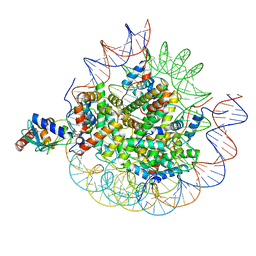 | | Cryo-EM structure of the human nucleosome core particle in complex with RNF168-UbcH5c | | Descriptor: | DNA (147-MER), E3 ubiquitin-protein ligase RNF168, Histone H2A type 1-B/E, ... | | Authors: | Hu, Q, Botuyan, M.V, Zhao, D, Cui, G, Mer, G. | | Deposit date: | 2023-10-22 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQ9
 
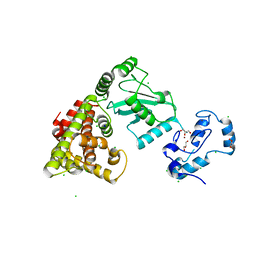 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 4-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, GLYCEROL, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8IHL
 
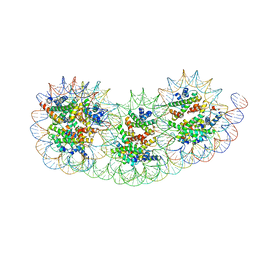 | | Overlapping tri-nucleosome | | Descriptor: | DNA (353-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Nishimura, M, Fujii, T, Tanaka, H, Maehara, K, Nozawa, K, Takizawa, Y, Ohkawa, Y, Kurumizaka, H. | | Deposit date: | 2023-02-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (7.64 Å) | | Cite: | Genome-wide mapping and cryo-EM structural analyses of the overlapping tri-nucleosome composed of hexasome-hexasome-octasome moieties.
Commun Biol, 7, 2024
|
|
8UQ8
 
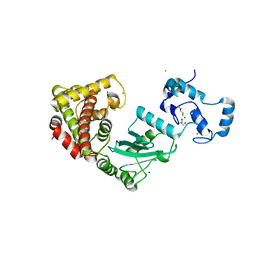 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 2-residue linker | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, GLYCEROL, ... | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQD
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (condition 2. RING not modeled in density) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3.893 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
8UQC
 
 | | Crystal structure of RNF168 (RING)-UbcH5c fused to H2A-H2B via a 20-residue linker (crystallization condition 2) | | Descriptor: | E3 ubiquitin-protein ligase RNF168,Ubiquitin-conjugating enzyme E2 D3,Histone H2B type 2-E,Histone H2A type 1-B/E, ZINC ION | | Authors: | Hu, Q, Botuyan, M.V, Mer, G. | | Deposit date: | 2023-10-23 | | Release date: | 2024-01-17 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.61 Å) | | Cite: | Mechanisms of RNF168 nucleosome recognition and ubiquitylation.
Mol.Cell, 84, 2024
|
|
7YBF
 
 | |
8WG5
 
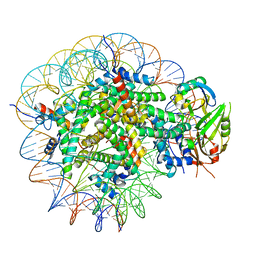 | | Cryo-EM structure of USP16 bound to H2AK119Ub nucleosome | | Descriptor: | DNA (147-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Ai, H.S, He, Z.Z, Deng, Z.H, Liu, L. | | Deposit date: | 2023-09-20 | | Release date: | 2023-12-27 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural and mechanistic basis for nucleosomal H2AK119 deubiquitination by single-subunit deubiquitinase USP16.
Nat.Struct.Mol.Biol., 2024
|
|
8CBN
 
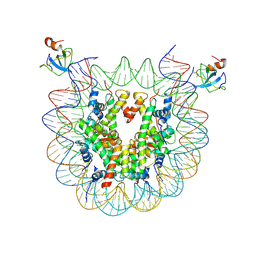 | | structure of LEDGF/p75 PWWP domain bound to the H3K36 trimethylated dinucleosome | | Descriptor: | Histone H2A, Histone H2B 1.1, Histone H3, ... | | Authors: | Koutna, E, Kouba, T, Novacek, J, Veverka, V. | | Deposit date: | 2023-01-25 | | Release date: | 2023-12-27 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Multivalency of nucleosome recognition by LEDGF.
Nucleic Acids Res., 51, 2023
|
|
7YRG
 
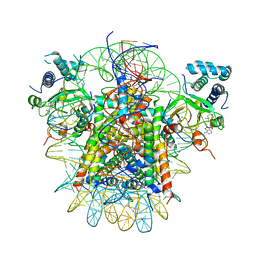 | | histone methyltransferase | | Descriptor: | DNA (146-MER), Histone H2A.Z, Histone H2B 1.1, ... | | Authors: | Li, H, Wang, W.Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-12-13 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural insight into H4K20 methylation on H2A.Z-nucleosome by SUV420H1.
Mol.Cell, 83, 2023
|
|
7XYF
 
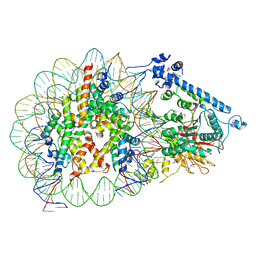 | |
7XYG
 
 | |
8JH2
 
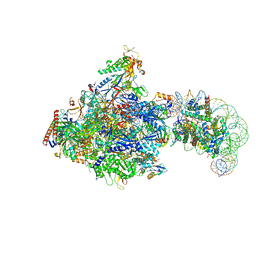 | | RNA polymerase II elongation complex bound with Elf1, Spt4/5 and foreign DNA, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (218-MER), DNA (40-MER), DNA-directed RNA polymerase subunit, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (5.7 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8JH4
 
 | | RNA polymerase II elongation complex containing 60 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8JH3
 
 | | RNA polymerase II elongation complex containing 40 bp upstream DNA loop, stalled at SHL(-1) of the nucleosome | | Descriptor: | DNA (198-MER), DNA-directed RNA polymerase subunit, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Akatsu, M, Fujita, R, Ogasawara, M, Ehara, H, Kujirai, T, Takizawa, Y, Sekine, S, Kurumizaka, H. | | Deposit date: | 2023-05-22 | | Release date: | 2023-11-29 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structures of RNA polymerase II-nucleosome complexes rewrapping transcribed DNA.
J.Biol.Chem., 299, 2023
|
|
8UX1
 
 | |
8JHG
 
 | | Native SUV420H1 bound to 167-bp nucleosome | | Descriptor: | DNA (160-MER), Histone H2A type 1-B/E, Histone H2B type 1-K, ... | | Authors: | Lin, F, Li, W. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-22 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.58 Å) | | Cite: | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
8JHF
 
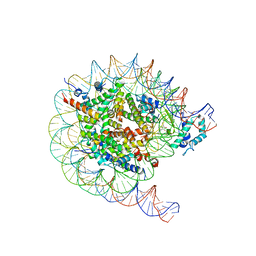 | | Native SUV420H1 bound to 167-bp nucleosome | | Descriptor: | DNA (160-MER), Histone H2A.Z, Histone H2B type 1-K, ... | | Authors: | Lin, F, Li, W. | | Deposit date: | 2023-05-23 | | Release date: | 2023-11-15 | | Last modified: | 2023-12-20 | | Method: | ELECTRON MICROSCOPY (3.68 Å) | | Cite: | Structural basis of nucleosomal H4K20 recognition and methylation by SUV420H1 methyltransferase.
Cell Discov, 9, 2023
|
|
7XFJ
 
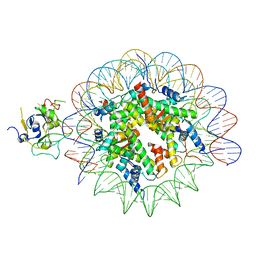 | | Structure of nucleosome-AAG complex (T-50I, post-catalytic state) | | Descriptor: | DNA (152-MER), DNA-3-methyladenine glycosylase, Histone H2A type 1, ... | | Authors: | Zheng, L, Tsai, B, Gao, N. | | Deposit date: | 2022-04-01 | | Release date: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural and mechanistic insights into the DNA glycosylase AAG-mediated base excision in nucleosome.
Cell Discov, 9, 2023
|
|
8SYP
 
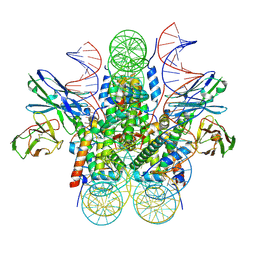 | | Genomic CX3CR1 nucleosome | | Descriptor: | DNA (162-MER), Histone H2A type 2-C, Histone H2B type 2-E, ... | | Authors: | Lian, T, Guan, R, Bai, Y. | | Deposit date: | 2023-05-25 | | Release date: | 2023-11-01 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Structural mechanism of synergistic targeting of the CX3CR1 nucleosome by PU.1 and C/EBP alpha.
Nat.Struct.Mol.Biol., 31, 2024
|
|
